应用化学 ›› 2023, Vol. 40 ›› Issue (11): 1558-1571.DOI: 10.19894/j.issn.1000-0518.230165
益肾排石方的指纹图谱及功效关联物质的作用机制
刘忠义1, 刘聪1, 马永璠1, 赵威1, 刘艳华2, 初洪波2( )
)
- 1.长春中医药大学药学院,长春 130117
2.长春中医药大学附属医院,长春 130021
-
收稿日期:2023-06-05接受日期:2023-10-08出版日期:2023-11-01发布日期:2023-12-01 -
通讯作者:初洪波 -
基金资助:吉林省自然科学基金(YDZJ202201ZYTS237);吉林省科技发展计划项目(20200404068YY)
Fingerprints and Action Mechanism of Efficacy-Associated Substances of Yishen Paishi Formula
Zhong-Yi LIU1, Cong LIU1, Yong-Fan MA1, Wei ZHAO1, Yan-Hua LIU2, Hong-Bo CHU2( )
)
- 1.School of Pharmaceutical Sciences Changchun University of Chinese Medicine,Changchun 130117,China
2.The Affiliated Hospital to Changchun University of Chinese Medicine,Changchun 130021,China
-
Received:2023-06-05Accepted:2023-10-08Published:2023-11-01Online:2023-12-01 -
Contact:Hong-Bo CHU -
About author:7377023@qq.com
-
Supported by:Jilin Provincial Natural Science Foundation(YDZJ202201ZYTS237);the Science and Technology Development Project of Jilin Province(20200404068YY)
摘要:
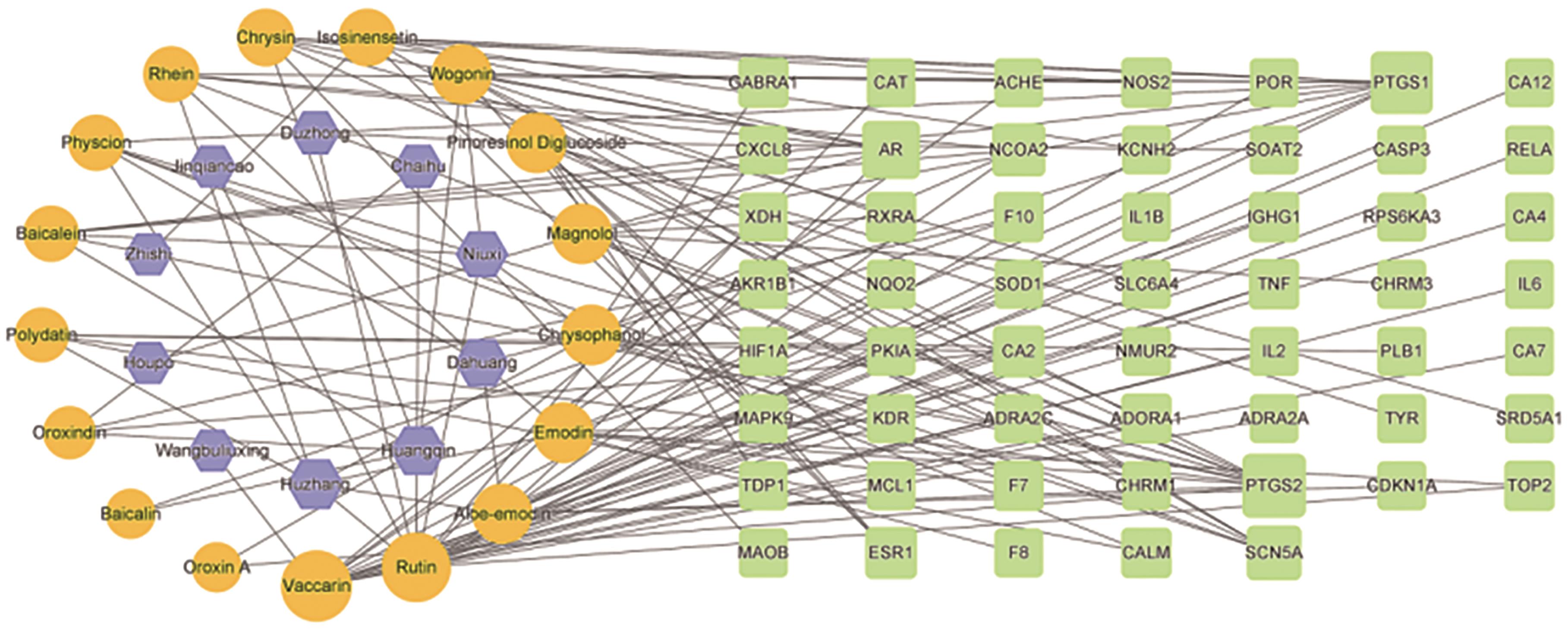
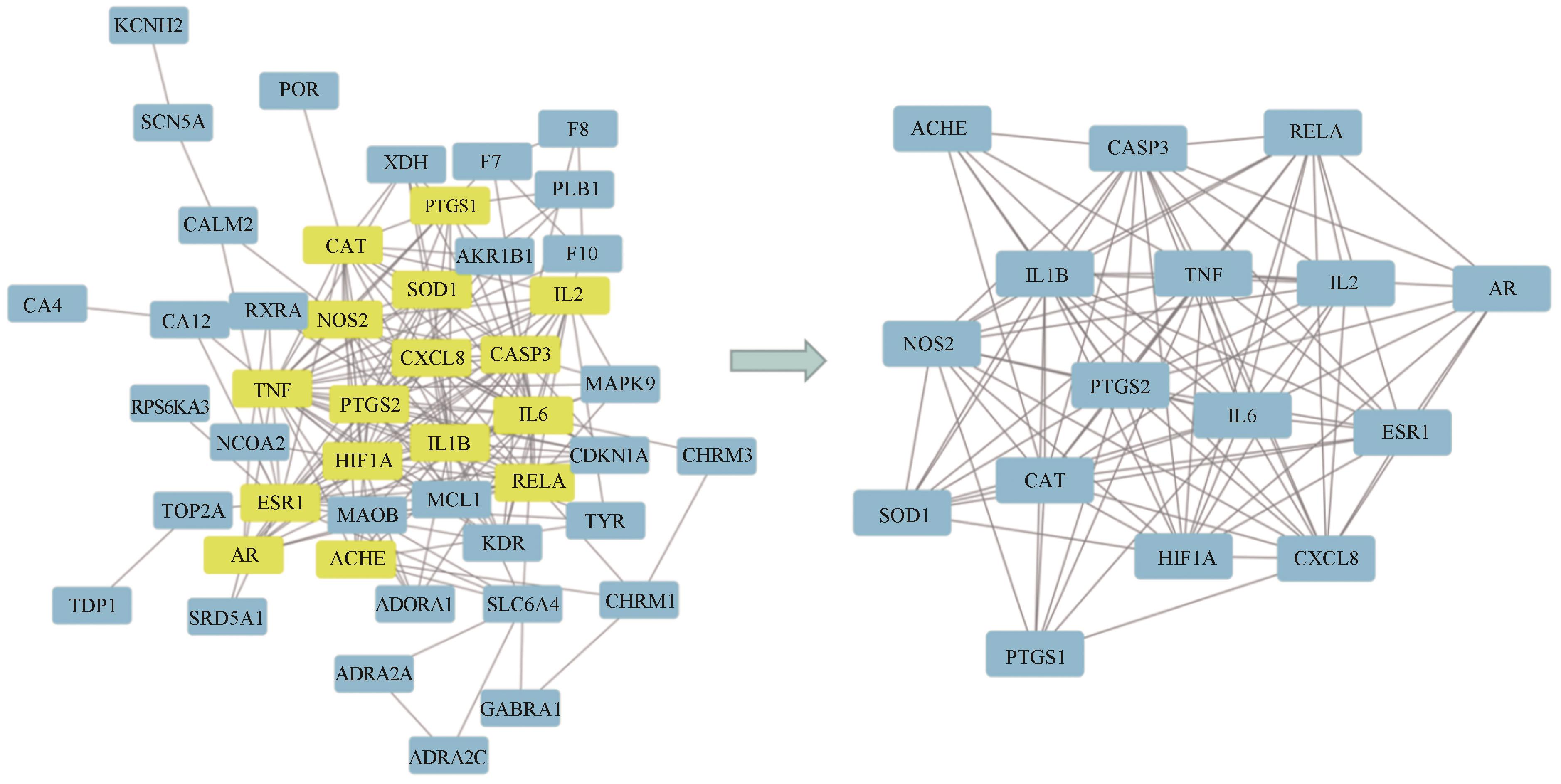
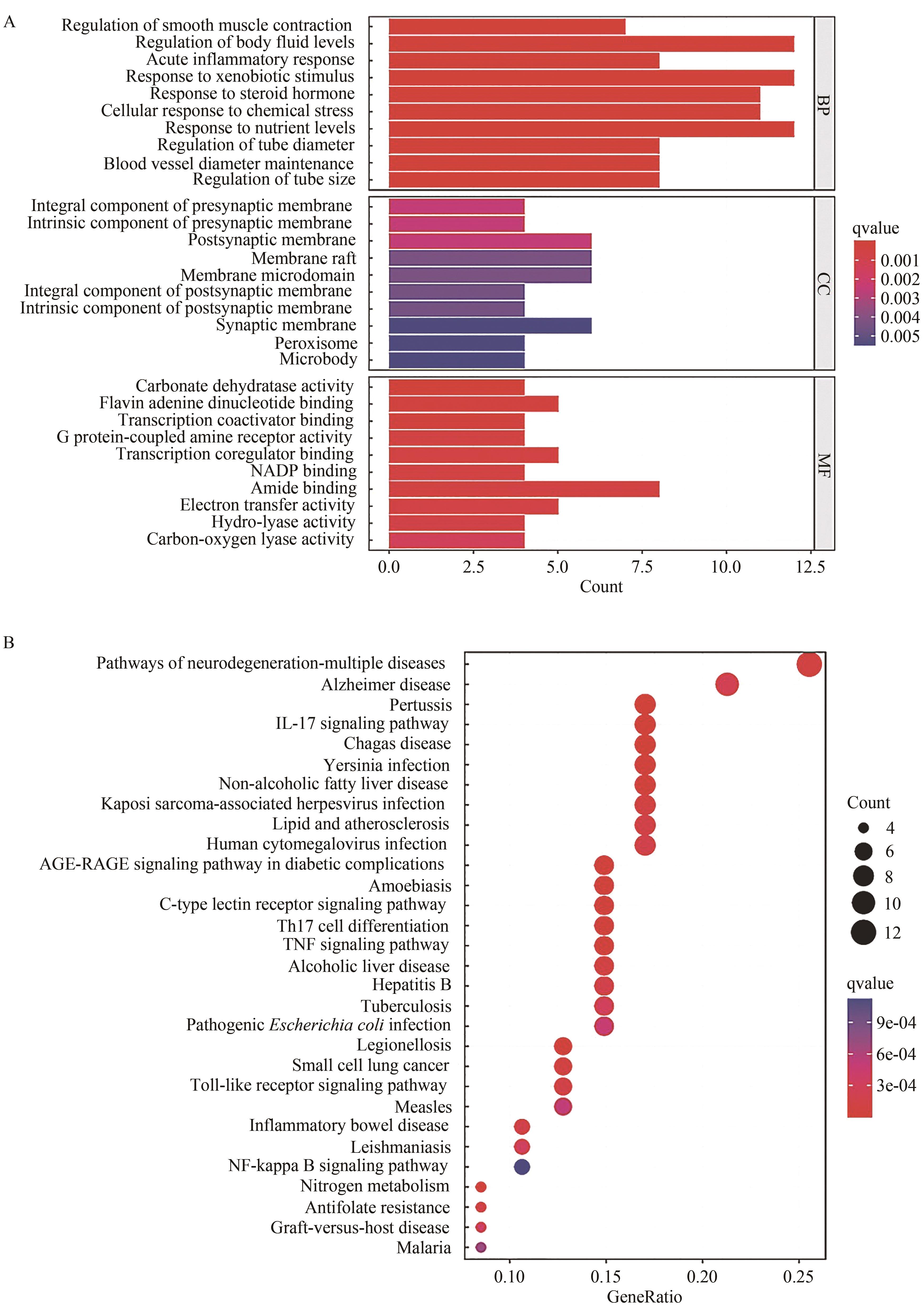
应用液质联用技术分析了益肾排石方中的化学成分,建立高效液相色谱法(HPLC)指纹图谱,结合生物信息学技术对关键成分、关键靶点和通路进行了预测分析。应用超高效液相串联质谱技术(UHPLC-MS/MS)对益肾排石方中的化学成分进行了鉴定。10批益肾排石方样品,应用HPLC建立指纹图谱,标记共有峰,进行共有峰归属,并进行相似度评价。应用生物信息学技术,基于指认出的成分,建立了“中药-成分-靶点”网络图,通过GEO数据库获得了肾结石相关的差异基因。对方中功效关联物质的作用机制进行了进一步的分析,并进行了分子对接验证。通过数据库匹配、元素组成和碎片结构分析,共从益肾排石方中鉴定出32种成分; 在对10批益肾排石方的指纹图谱的建立过程中,通过对照品指认确定了王不留行黄酮苷、松脂醇二葡萄糖苷、虎杖苷、黄芩苷、黄芩素、汉黄芩苷、芦丁、木蝴蝶苷A、大黄酸、异橙黄酮、芦荟大黄素、白杨素、大黄酚、厚朴酚、大黄素甲醚、汉黄芩素和大黄素17个共有峰,10批益肾排石方相似度结果均在合理范围内,且17个共有峰均能归属到虎杖、黄芩、牛膝、柴胡、金钱草、枳实、大黄、厚朴、杜仲和王不留行10味中药。结合生物信息学筛选了ESR1和PTGS1为关键基因,与17种药效成分进行分子对接验证,预测大黄酸和王不留行黄酮苷为关键成分。
中图分类号:
引用本文
刘忠义, 刘聪, 马永璠, 赵威, 刘艳华, 初洪波. 益肾排石方的指纹图谱及功效关联物质的作用机制[J]. 应用化学, 2023, 40(11): 1558-1571.
Zhong-Yi LIU, Cong LIU, Yong-Fan MA, Wei ZHAO, Yan-Hua LIU, Hong-Bo CHU. Fingerprints and Action Mechanism of Efficacy-Associated Substances of Yishen Paishi Formula[J]. Chinese Journal of Applied Chemistry, 2023, 40(11): 1558-1571.
| Time/min | Mobile phase B/% | Mobile phase A/% | Other conditions |
|---|---|---|---|
| 0 | 5 | 95 | Column temperature: 30 ℃ Flow rates: 0.3 mL/min Sample size: 2 μL |
| 2 | 5 | 95 | |
| 6 | 30 | 70 | |
| 7 | 30 | 70 | |
| 12 | 78 | 22 | |
| 14 | 78 | 22 | |
| 17 | 95 | 5 | |
| 20 | 95 | 5 | |
| 21 | 95 | 5 | |
| 25 | 5 | 95 |
表1 超高效液相色谱梯度洗脱程序
Table 1 Gradient elution procedure of ultra performance liquid chromatography
| Time/min | Mobile phase B/% | Mobile phase A/% | Other conditions |
|---|---|---|---|
| 0 | 5 | 95 | Column temperature: 30 ℃ Flow rates: 0.3 mL/min Sample size: 2 μL |
| 2 | 5 | 95 | |
| 6 | 30 | 70 | |
| 7 | 30 | 70 | |
| 12 | 78 | 22 | |
| 14 | 78 | 22 | |
| 17 | 95 | 5 | |
| 20 | 95 | 5 | |
| 21 | 95 | 5 | |
| 25 | 5 | 95 |
| Time/min | Mobile phase B/% | Mobile phase A/% | Other conditions |
|---|---|---|---|
| 0 | 5 | 95 | Column temperature: 25 ℃ Flow rates: 1 mL/min Sample size: 10 μL Detection wavelength: 254 nm |
| 10 | 5 | 95 | |
| 25 | 10 | 90 | |
| 40 | 15 | 85 | |
| 60 | 20 | 80 | |
| 70 | 30 | 70 | |
| 80 | 40 | 60 | |
| 90 | 50 | 50 | |
| 100 | 60 | 40 | |
| 110 | 70 | 30 | |
| 120 | 80 | 20 | |
| 130 | 5 | 95 | |
| 140 | 5 | 95 |
表2 高效液相色谱梯度洗脱程序
Table 2 Gradient elution program of high performance liquid chromatography
| Time/min | Mobile phase B/% | Mobile phase A/% | Other conditions |
|---|---|---|---|
| 0 | 5 | 95 | Column temperature: 25 ℃ Flow rates: 1 mL/min Sample size: 10 μL Detection wavelength: 254 nm |
| 10 | 5 | 95 | |
| 25 | 10 | 90 | |
| 40 | 15 | 85 | |
| 60 | 20 | 80 | |
| 70 | 30 | 70 | |
| 80 | 40 | 60 | |
| 90 | 50 | 50 | |
| 100 | 60 | 40 | |
| 110 | 70 | 30 | |
| 120 | 80 | 20 | |
| 130 | 5 | 95 | |
| 140 | 5 | 95 |

图1 混合对照品(A)、益肾排石供试品(B)和10批益肾排石方(C)的HPLC色谱图S1-S10. 10 batches of kidney-enhancing and stone-dispelling formulae charts; S11. Control charts; R. Mixed Standards Chart 1.Pinoresinol diglucoside; 2.Vaccarin; 3.Polydatin; 4.Rutinum; 5.Oroxin A; 6.Baicalin; 7.Wogonoside; 8.Baicalein; 9.Isosinensetin; 10.Aloe-emodin; 11.Rhein; 12.Wogonin; 13.Chrysin; 14.Emodin; 15.Magnolol; 16.Chrysophanol; 17.Physcion
Fig.1 HPLC chromatogram of mixed reference substance (A), Yishen Paishi test substance (B) and 10 batches of Yishen Paishi prescription (C)
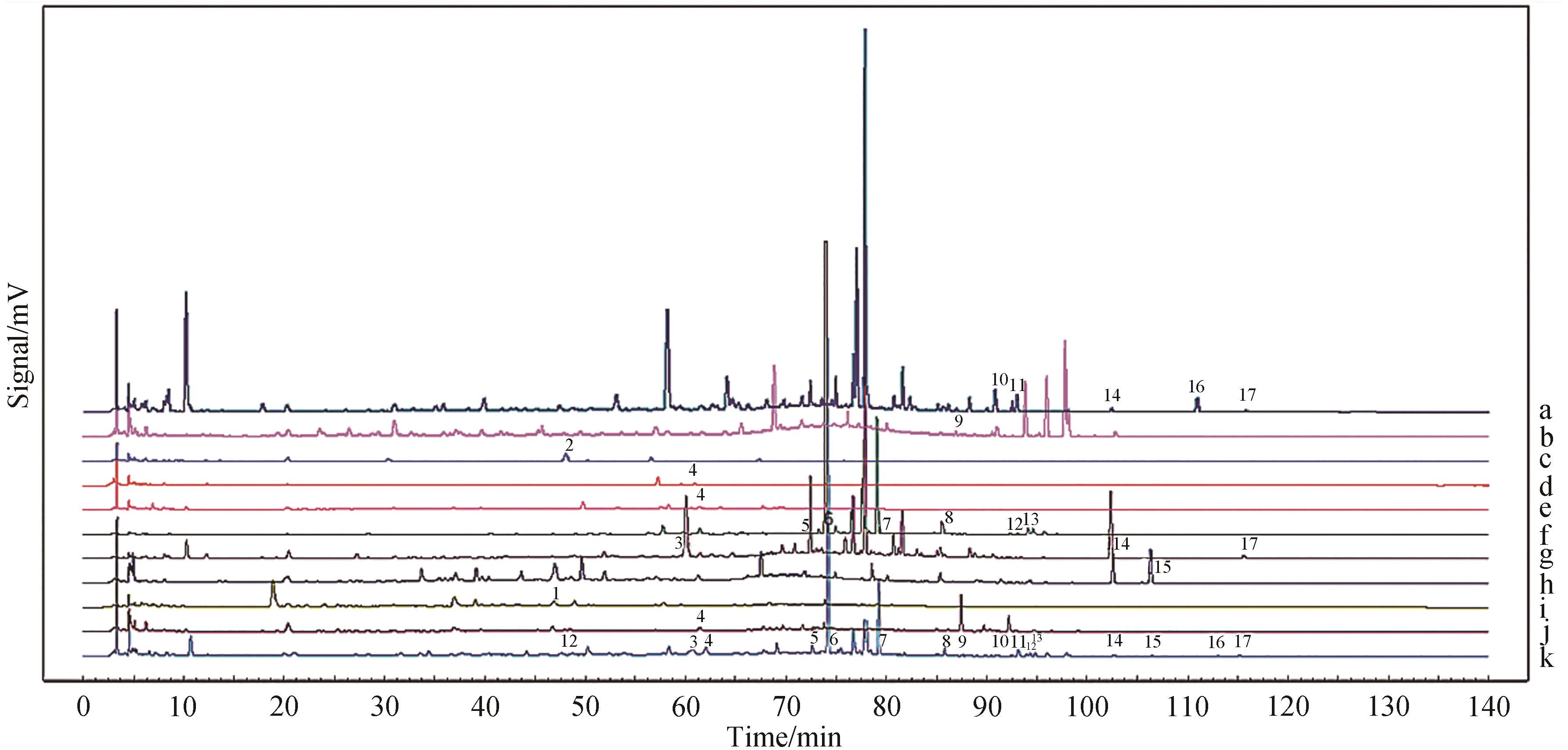
图2 益肾排石单味药色谱图a.Rheum; b.Citrus; c.Vaccariae Semen; d.Achyranthes bidentata; e.Lysimachia christinae Hance; f.Scutellaria baicalensis Georgi; g.Polygonum cuspidatum; h.Magnolia officinalis; i.Eucommia ulmoides; j.Bupleurum; k.YSPS
Fig.2 Chromatogram of a single drug for Yishen Paishi

图7 肾结石疾病相关差异基因分析结果A. Box diagram (used to view the correction of the sample); B. PCA analysis diagram of chip data set GSE73680 (used to view clustering among sample groups); C. Volcano map of GSE73680 differentially expressed gene; D. Heat map of GSE73680 differential gene cluster analysis
Fig.7 Analysis results of differential genes associated with kidney stone disease
| Filter conditions | Quantities |
|---|---|
| |log FC|>2 &P<0.05 | 325 |
| |log FC|>1 & P<0.05 | 6 227 |
| |log FC|>0.58 & P<0.05 | 12 316 |
表3 差异基因筛选结果表
Table 3 Screening results of differential genes
| Filter conditions | Quantities |
|---|---|
| |log FC|>2 &P<0.05 | 325 |
| |log FC|>1 & P<0.05 | 6 227 |
| |log FC|>0.58 & P<0.05 | 12 316 |
| Target-Active ingredients | Affinity/(kJ·mol-1) | Target-Active ingredients | Affinity/(kJ·mol-1) |
|---|---|---|---|
| ESR1-rhein | -38.49 | PTGS1-Vaccarin | -40.58 |
| ESR1-chrysin | -38.07 | PTGS1-Rutinum | -40.17 |
| ESR1-baicalein | -38.07 | PTGS1-chrysin | -38.91 |
| ESR1-emodin | -38.07 | PTGS1-rhein | -38.49 |
| ESR1-aloe-emodin | -37.24 | PTGS1-baicalein | -38.07 |
| ESR1-Chrysophanol | -37.24 | PTGS1-Wogonoside | -38.07 |
| ESR1-Vaccarin | -35.98 | PTGS1-Oroxin A | -37.66 |
| ESR1-Wogonin | -35.56 | PTGS1-Baicalin | -37.66 |
| ESR1-physcion | -35.15 | PTGS1-aloe-emodin | -37.24 |
| ESR1-Oroxin A | -33.89 | PTGS1-Chrysophanol | -37.24 |
| ESR1-Rutinum | -33.89 | PTGS1-physcion | -36.40 |
| ESR1-Baicalin | -33.47 | PTGS1-emodin | -35.98 |
| ESR1-polydatin | -33.05 | PTGS1-polydatin | -36.40 |
| ESR1-pinoresinol diglucoside | -32.22 | PTGS1-pinoresinol diglucoside | -34.73 |
| ESR1-Wogonoside | -31.80 | PTGS1-Wogonin | -34.31 |
| ESR1-Magnolol | -30.96 | PTGS1-Isosinensetin | -34.31 |
| ESR1-Isosinensetin | -28.87 | PTGS1-Magnolol | -30.96 |
表4 核心靶点与活性成分对接的结合最小自由能
Table 4 Minimum free energy of binding between core target and active ingredient
| Target-Active ingredients | Affinity/(kJ·mol-1) | Target-Active ingredients | Affinity/(kJ·mol-1) |
|---|---|---|---|
| ESR1-rhein | -38.49 | PTGS1-Vaccarin | -40.58 |
| ESR1-chrysin | -38.07 | PTGS1-Rutinum | -40.17 |
| ESR1-baicalein | -38.07 | PTGS1-chrysin | -38.91 |
| ESR1-emodin | -38.07 | PTGS1-rhein | -38.49 |
| ESR1-aloe-emodin | -37.24 | PTGS1-baicalein | -38.07 |
| ESR1-Chrysophanol | -37.24 | PTGS1-Wogonoside | -38.07 |
| ESR1-Vaccarin | -35.98 | PTGS1-Oroxin A | -37.66 |
| ESR1-Wogonin | -35.56 | PTGS1-Baicalin | -37.66 |
| ESR1-physcion | -35.15 | PTGS1-aloe-emodin | -37.24 |
| ESR1-Oroxin A | -33.89 | PTGS1-Chrysophanol | -37.24 |
| ESR1-Rutinum | -33.89 | PTGS1-physcion | -36.40 |
| ESR1-Baicalin | -33.47 | PTGS1-emodin | -35.98 |
| ESR1-polydatin | -33.05 | PTGS1-polydatin | -36.40 |
| ESR1-pinoresinol diglucoside | -32.22 | PTGS1-pinoresinol diglucoside | -34.73 |
| ESR1-Wogonoside | -31.80 | PTGS1-Wogonin | -34.31 |
| ESR1-Magnolol | -30.96 | PTGS1-Isosinensetin | -34.31 |
| ESR1-Isosinensetin | -28.87 | PTGS1-Magnolol | -30.96 |
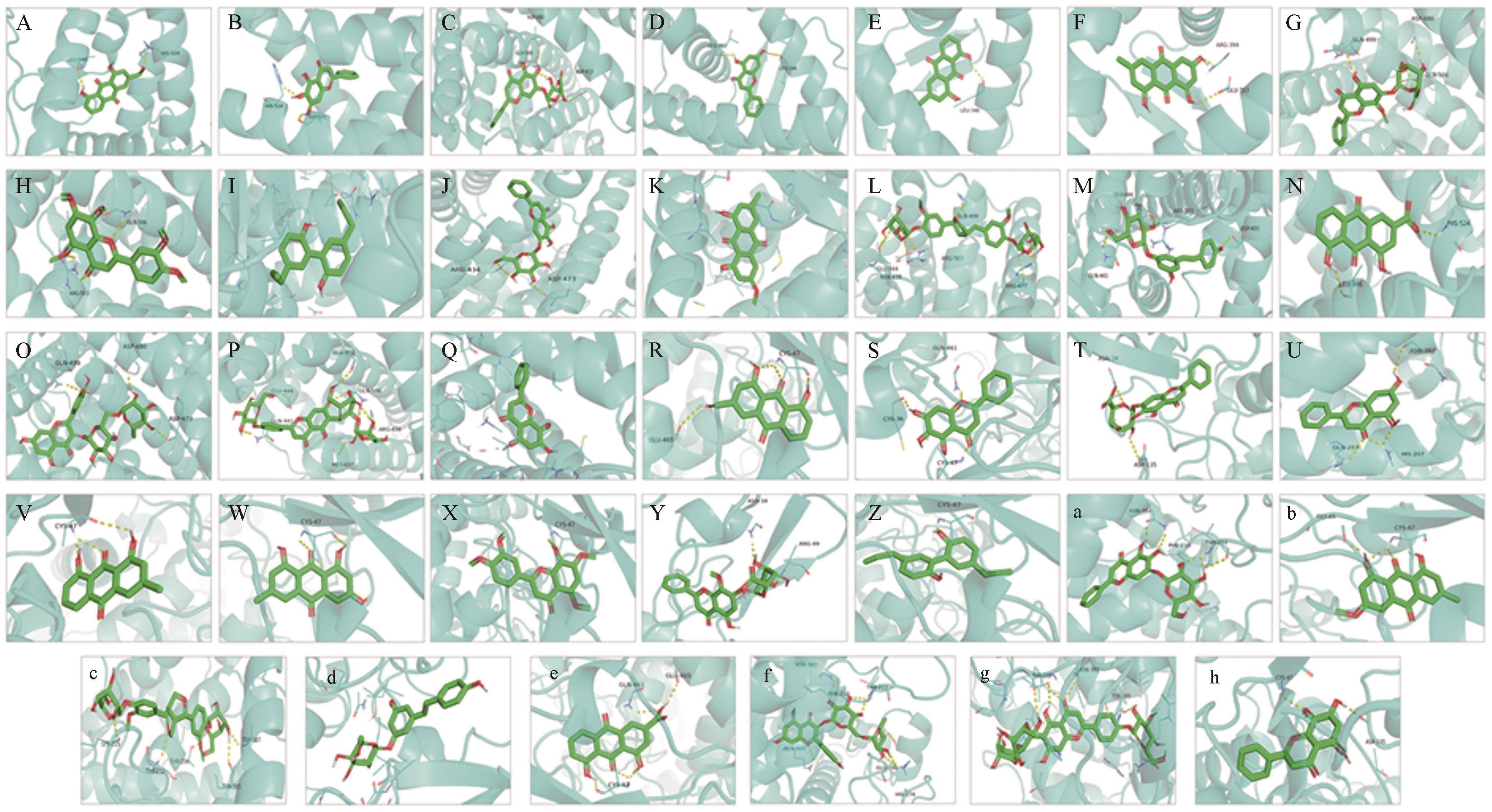
图9 活性成分与关键基因的分子对接3D图A: ESR1 and aloe-emodin; B: ESR1 and baicalein; C: ESR1 and Baicalin; D: ESR1 and chrysin; E: ESR1 and Chrysophanol; F: ESR1 and emodin; G: ESR1 and Wogonoside; H: ESR1 and Isosinensetin; I: ESR1 and Magnolol; J: ESR1 and Oroxin A; K: ESR1 and physcion; L: ESR1 and pinoresinol diglucoside; M: ESR1 and polydatin; N: ESR1 and rhein; O: ESR1 and Rutinum; P: ESR1 and Vaccarin; Q: ESR1 and Wogonin; R: PTGS1 and aloe-emodin; S: PTGS1 and baicalein; T: PTGS1 and Baicalin; U: PTGS1 and chrysin; V: PTGS1 and Chrysophano; W: PTGS1 and emodin; X: PTGS1 and Isosinensetin; Y: PTGS1 and Wogonoside; Z: PTGS1 and Magnolol; a: PTGS1 and Oroxin A; b: PTGS1 and physcion; c: PTGS1 and pinoresinol diglucoside; d: PTGS1 and polydatin; e: PTGS1 and rhein; f: PTGS1 and Rutinum; g: PTGS1 and Vaccarin; h: PTGS1 and Wogonin
Fig.9 3D diagram of molecular docking between active ingredients and key genes
| 1 | 石娅, 刘文, 刘兴德, 等. 基于PI3K/Akt信号通路探究当归补血汤干预大鼠实验性脑缺血再灌注损伤的作用机制[J]. 中草药, 2022, 53(16): 5052-5065. |
| SHI Y, LIU W, LIU X D, et al. Mechanism of Danggui Buxue decoction in intervention of experimental cerebral ischemia reperfusion injury in rats based on PI3K/Akt signaling pathway[J]. Chin Tradit Herb Drugs, 2022, 53(16): 5052-5065. | |
| 2 | 涂宛晴, 梁秋儿, 谢鹏程, 等. 基于TNF-α/IKKβ/NF-κB信号通路研究消积丸治疗ApoE-/-小鼠动脉粥样硬化的机制[J].中草药, 2022, 53(16): 5074-5084. |
| TU W Q, LIANG Q E, XIE P C, et al. Mechanism of Xiaoji Pills in treatment of atherosclerosis in ApoE-∕- mice based on TNF-α/IKKβ/NF-κB signaling pathway[J]. Chin Tradit Herb Drugs, 2022, 53(16): 5074-5084. | |
| 3 | 牛明, 张斯琴, 张博, 等. 《网络药理学评价方法指南》解读[J]. 中草药, 2021, 52(14): 4119-4129. |
| NIU M, ZHANG S Q, ZHANG B, et al. Interpretation of network pharmacology evaluation method guidance[J]. Chin Tradit Herb Drugs, 2021, 52(14): 4119-4129. | |
| 4 | 臧凝子, 李品, 庞立健, 等. 基于网络药理学和生物信息学筛选小青龙汤治疗支气管哮喘的关键基因和通路[J]. 中国实验方剂学杂志, 2021, 27(3): 171-183. |
| ZANG N Z, LI P, PANG L J, et al. Exploring key genes and signaling pathways in treatment of bronchial asthma with Xiao Qinglongtang based on network pharmacology and bioinformatics[J]. Chin J Exp Tradit Med Formulae, 2021, 27(3): 171-183. | |
| 5 | LIU J Q, LIU J, TONG X L, et al. Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of Huai Hua San against ulcerative colitis[J]. Drug Des Dev Ther, 2021, 15: 15: 3255-3276. |
| 6 | 姚令文, 刘燕, 郑笑为, 等. 指纹图谱、特征图谱技术在中药材和中成药中的应用[J]. 中国新药杂志, 2018, 27(8): 934-939. |
| YAO L W, LIU Y, ZGENG X W, et al. Application of the fingerprint and characteristic fingerprint technology in traditional Chinese medicinal materials and Chinese patent medicine[J]. Chin J New Drugs, 2018, 27(8): 934-939. | |
| 7 | 汪翔, 夏启东, 寻阳. 基于网络药理学和分子对接分析探讨五苓散治疗肾结石的潜在作用机制[J]. 广州医科大学学报, 2022, 50(5): 42-46. |
| WANG X, XIA Q D, XUN Y. Network pharmacological study and molecular docking analysis on potential mechanism of Wuling powder in treatment of kidney stone[J]. Acad J Guangzhou Med Univ, 2022, 50(5): 42-46. | |
| 8 | KIM A R, JUNG M C, JEONG H I, et al. Antioxidative and cellular protective effects of lysimachia christinae hance extract and fractions[J]. Appl Chem Eng, 2018, 29(2): 176-184. |
| 9 | KIRINO A, TAKASUKA Y, NISHI A, et al. Analysis and functionality of major polyphenolic components of polygonum cuspidatum (itadori)[J]. J Nutr Sci Vitaminol, 2012, 58(4): 278-286. |
| 10 | ZHANG H X, LIU M C. Separation procedures for the pharmacologically active components of rhubarb[J]. J Chromatogr B, 2004, 812(1/2): 175-181. |
| 11 | COSSIGNANI L, BLASI F, SIMONETTI M, et al. Fatty acids and phytosterols to discriminate geographic origin of Lycium barbarum berry[J]. Food Anal Methods, 2018, 11: 1180-1188. |
| 12 | HAN H K, VAN ANH L T. Modulation of P-glycoprotein expression by honokiol, magnolol and 4-O-methylhonokiol, the bioactive components of Magnolia officinalis[J]. Anticancer Res, 2012, 32(10): 4445-4452. |
| 13 | CHEUNG H Y, LAI W P, CHEUNG M S, et al. Rapid and simultaneous analysis of some bioactive components in Eucommia ulmoides by capillary electrophoresis[J]. J Chromatogr A, 2003, 989(2): 303-310. |
| 14 | LI X M, LIU Y, WANG S Y, et al. Identification and potential mechanism of different components from the aerial part of Bupleurum chinense DC. for epileptic treatment[J]. Nat Prod Res, 2022, 36(23): 6137-6142. |
| 15 | ZHANG J, CHOU G X, LIU Z J, et al. In vitro cytotoxicity and antioxidation of a whole fruit extract of Liquidambar formosana exerted by different constituents[J]. Eur J Med Plants, 2015, 6(1): 34. |
| 16 | 罗思妮, 彭致铖, 范倩, 等. 经典名方小承气汤中化学成分的UPLC-Q-Orbitrap-MS分析[J]. 中国实验方剂学杂志, 2021, 27(23): 1-10. |
| LUO S N, PENG Z C, FAN Q, et al. Analysis on chemical constituents in Xiao Chengqitang by UPLC-Q-Orbitrap-MS[J]. Chin J Exp Tradit Med Formulae, 2021, 27(23): 1-10. | |
| 17 | SMOOT M E, ONO K, RUSCHEINSKI J, et al. Cytoscape 2.8: new features for data integration and network visualization[J]. Bioinformatics, 2011, 27(3): 431-432. |
| 18 | 宋晓彤, 季旭明, 于华芸, 等. 小承气汤“成分-靶点-通路”的网络药理学研究[J]. 世界科学技术-中医药现代化, 2020, 22(9): 3150-3160. |
| SONG X T, JI X M, YU H Y, et al. Network pharmacology study of “ingredients-targets-pathways” of Xiaochengqi decoction[J]. Mod Traditional Chin Med Mater Med Would Sci Technol, 2020, 22(9): 3150-3160. | |
| 19 | MERING C V, HUYNEN M, JAEGGI D, et al. STRING: a database of predicted functional associations between proteins[J]. Nucleic Acids Res, 2003, 31(1): 258-261. |
| 20 | ZHANG J, XING Z H, MA M M, et al. Gene ontology and KEGG enrichment analyses of genes related to age-related macular degeneration[J]. BioMed Res Int, 2014, 2014: 450386. |
| 21 | 刘本涛, 袁彩英, 陈睿, 等. 基于GEO数据库及网络药理学研究绞股蓝治疗冠心病的作用机制[J]. 中成药, 2021, 43(10): 2885-2892. |
| LIU B T, YUAN C Y, CHEN R, et al. Mechanism of Gynostemma pentaphyllum in treating coronary heart disease based on GEO database and network pharmacology[J]. Chin Tradit Pat Med, 2021, 43(10): 2885-2892. | |
| 22 | 赵丽君, 马艳苗, 梁凯, 等. 基于芯片分析联合网络药理学探寻淫羊藿干预乳腺癌干细胞的生物标志物及靶点机制[J]. 中国实验方剂学杂志, 2021, 27(11): 195-204. |
| ZHAO L J, MA Y M, LIANG K, et al. Biomarkers and target mechanisms of epimedii folium intervention on breast cancer stem cells based on chip analysis and network pharmacology[J]. Chin J Exp Tradit Med Formulae, 2021, 27(11): 195-204. | |
| 23 | HSIN K Y, GHOSH S, KITANO H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology[J]. PLoS One, 2018, 8(12): e83922. |
| 24 | 刘海朝, 曹敏, 解露露, 等. 雌激素抑制肾草酸钙结石形成机制的研究进展[J]. 临床泌尿外科杂志, 2021, 36(1): 72-75. |
| LIU H Z, CAO M, XIE L L, et al. Research progress on the mechanism of estrogen inhibiting the formation of renal calcium oxalate stones[J]. J Clin Urol, 2021, 36(1): 72-75. | |
| 25 | 刘海朝. 基于网络药理学补肾化石汤抗肾结石形成的机制研究[D]. 天津: 天津中医药大学, 2021. |
| LIU H Z. Study on the mechanism of Bushen Huashi Decoction against renal calculi formation based on network pharmacology[D]. Tianjin: Tianjin University of Traditional Chinese Medicine, 2021. | |
| 26 | BOURGONJE A R, ABDULLE A E, AL-RAWAS A M, et al. Systemic oxidative stress is increased in postmenopausal women and independently associates with homocysteine levels[J]. Int J Mol Sci, 2020, 21(1): 314. |
| 27 | MARCUS R, MADVIG P, CRIM M, et al. Conjugated estrogens in the treatment of postmenopausal women with hyperparathyroidism[J]. Ann Intern Med, 1984, 100(5): 633-640. |
| 28 | DOSHI S B, AGARWAL A. The role of oxidative stress in menopause[J]. J Food Sci, 2013, 4(3): 140. |
| 29 | STUBBINS R E, NAJJAR K, HOLCOMB V B, et al. Oestrogen alters adipocyte biology and protects female mice from adipocyte inflammation and insulin resistance[J]. Diabetes Obes Metab, 2012, 14(1): 58-66. |
| 30 | MOOSMANN B, BEHL C. The antioxidant neuroprotective effects of estrogens and phenolic compounds are independent from their estrogenic properties[J]. Proc Natl Acad Sci, 1999, 96(16): 8867-8872. |
| 31 | KRAMER H J M, GRODSTEIN F, STAMPFER M J, et al. Menopause and postmenopausal hormone use and risk of incident kidney stones[J]. J Am Soc Nephrol, 2003, 14(5): 1272-1277. |
| 32 | MAALOUF N M, SATO A H, WELCH B J, et al. Postmenopausal hormone use and the risk of nephrolithiasis: results from the Women's Health Initiative hormone therapy trials[J]. Arch Intern Med, 2010, 170(18): 1678-1685. |
| 33 | TURNER R J, KERBER I J. Renal stones, timing hypothesis, and eu-estrogenemia[J]. Menopause, 2012, 19(1): 104-108. |
| 34 | ASSIMOS D G. Re: activities of calcium-related ion channels during the formation of kidney stones in an infection-induced urolithiasis rat model[J]. J Urol, 2020, 203(2): 246-247. |
| 35 | TAGUCHI K, HAMAMOTO S, OKADA A, et al. Genome-wide gene expression profiling of Randall′s plaques in calcium oxalate stone formers[J]. J Am Soc Nephrol, 2017, 28(1): 333-347. |
| 36 | PARK S J, KIM J H, HA T S, et al. The role of Interleukin-23/Interleukin-17 axis in coexisting anti-glomerular basement membrane disease and lupus nephritis[J]. Saudi J Kidney Dis Transpl, 2013, 24(3): 596. |
| 37 | 王兴玥, 高冷, 戚良晨, 等. 基于网络药理学及分子对接技术研究向海雁鹅血的抗肺癌活性成分[J]. 应用化学, 2022, 39(11): 1680-1692. |
| WANG X Y, GAO L, QI L C, et al. Study on anti-lung cancer active ingredients from blood of Xianghai wild goose based on network pharmacology and molecular docking technology[J]. Chin J Appl Chem, 2022, 39(11): 1680-1692. |
| [1] | 冷泽山, 郭洪梅, 蔡函青, 张剑. 高效液相色谱-电雾式检测器法同时检测食品中8种人工甜味剂的应用[J]. 应用化学, 2023, 40(3): 436-440. |
| [2] | 李玉凰, 卢泽毅, 袁红梅, 王刚, 张成江. 酰腙键聚合物凝胶的制备及其在硝基呋喃类药物分析中的应用[J]. 应用化学, 2023, 40(1): 100-108. |
| [3] | 付豪, 熊婉淇, 王祯, 段爱红, 袁黎明. 手性樟脑衍生的共价有机骨架材料用作高效液相色谱固定相分离手性化合物[J]. 应用化学, 2022, 39(9): 1371-1381. |
| [4] | 李淑芳, 郝学飞, 王红旗, 郭晓萌, 冯书惠, 尹海燕, 刘冬梅, 于永杰. 基于超高效液相色谱联用高分辨质谱自动解析方法的金银花不同加工方式分析[J]. 应用化学, 2022, 39(5): 819-827. |
| [5] | 孙慧婧, 崔冬妮. 固相萃取-同位素稀释-超高效液相色谱-串联质谱法测定水中涕灭威、涕灭威亚砜和涕灭威砜[J]. 应用化学, 2022, 39(3): 470-479. |
| [6] | 陈瑶, 唐英. 柱前衍生化高效液相色谱法测定氯氮平中肼含量[J]. 应用化学, 2022, 39(02): 322-331. |
| [7] | 霍颖异, 谢木西丁·买热帕提, 吴敏. 超高效液相色谱-质谱联用法同时快速测定细胞中的4种吡啶核苷酸辅酶[J]. 应用化学, 2022, 39(02): 332-339. |
| [8] | 朱富强, 丁卫平, 韩岩君, 田洪根. 脂质去除分散固相萃取-超高效液相色谱-串联质谱法测定动物源性食品中5种α2-受体激动剂[J]. 应用化学, 2021, 38(6): 713-721. |
| [9] | 张娜, 李乐乐, 黄鑫, 刘淑莹. 超高效液相色谱-三重四极杆质谱联用结合固相甲基化技术测定不同生长环境人参中寡糖分布[J]. 应用化学, 2021, 38(3): 247-255. |
| [10] | 李乐, 谭璐瑩, 王彩霞, 李坤, 李平亚, 刘金平, 刘云鹤. 超高效液相色谱-四极杆飞行时间质谱法鉴定西洋参果梗化学成分[J]. 应用化学, 2021, 38(3): 256-270. |
| [11] | 闫伊萌, 岳可心, 刘玉生, 陈革, 田涵雯, 刘忠英, 刘志强, 宋凤瑞, 皮子凤. 基于超高效液相色谱-四极杆-飞行时间串联质谱联用技术的黄英咳喘糖浆化学成分分析[J]. 应用化学, 2021, 38(3): 276-288. |
| [12] | 王聪, 赵晓宇, 王海燕, 曹进, 王钢力. 高效液相色谱-三重四极杆质谱法测定动物肌肉中73种兽药残留[J]. 应用化学, 2021, 38(12): 1663-1675. |
| [13] | 王为, 吴彦, 楚刘喜, 袁琳, 朱敏惠, 杨瑾, 邓慧华. HIV感染者头发中糖皮质激素和内源性大麻素的高效液相-串联质谱检测[J]. 应用化学, 2021, 38(11): 1521-1530. |
| [14] | 汪海涛, 刘海锋, 刘艳, 赵文博, 高润利, 段晶钟, 张远鹏. 多种分析方法测定邻氯代苯亚甲基丙二腈的纯度[J]. 应用化学, 2020, 37(7): 810-815. |
| [15] | 胡学一, 胡益涛, 方云, 夏咏梅, 钱飞. 正相高效液相色谱法分析壬基环己醇聚氧乙烯醚中残留壬基环己醇[J]. 应用化学, 2020, 37(7): 816-822. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||