
应用化学 ›› 2022, Vol. 39 ›› Issue (1): 3-17.DOI: 10.19894/j.issn.1000-0518.210479
生成模型在蛋白质序列设计中的应用
伍青林1, 任玉彬2, 翟小威1, 陈东1( ), 刘凯2(
), 刘凯2( )
)
- 1.浙江大学能源工程学院,杭州 310012
2.清华大学化学系,北京 100084
-
收稿日期:2021-09-26接受日期:2021-11-11出版日期:2022-01-01发布日期:2022-01-10 -
通讯作者:陈东,刘凯 -
基金资助:国家自然科学基金(21878258┫浙江省自然科学基金┣Y20B060027);资助
Protein Sequence Design Using Generative Models
WU Qing-Lin1, REN Yu-Bin2, ZHAI Xiao-Wei1, CHEN Dong1( ), LIU Kai2(
), LIU Kai2( )
)
- 1.College of Energy Engineering,Zhejiang University,Hangzhou 310012,China
2.Department of Chemistry,Tsinghua University,Beijing 100084,China
-
Received:2021-09-26Accepted:2021-11-11Published:2022-01-01Online:2022-01-10 -
Contact:Dong CHEN,Kai LIU -
About author:kailiu@mail.tsinghua.edu.cn
kailiu@mail.tsinghua.edu.cn
-
Supported by:the National Natural Science Foundation of China(21878258);Zhejiang Provincial Natural Science Foundation of China(Y20B060027)
摘要:
蛋白质是一切生命体的物质基础,是生命活动的主要承担者,参与各种生理功能的调节。设计具有特定功能的蛋白质在蛋白质工程、生物医药、材料科学等领域具有重要意义。蛋白质序列设计的目标是设计能够折叠成期望结构并具有相应功能的氨基酸序列,是所有理性蛋白质工程的核心问题,具有极其重要的研究和应用潜力。随着蛋白质序列数据的指数型增长和深度学习技术的快速发展,生成模型越来越多地被应用于蛋白质序列设计。本文简要介绍了蛋白质序列设计的重要意义和主要方法,概述了应用于蛋白质序列设计的主要生成模型,介绍了近年来生成模型在蛋白质序列表示、生成和优化方面的最新研究和应用现状,并对未来的发展方向进行讨论与展望。
中图分类号:
引用本文
伍青林, 任玉彬, 翟小威, 陈东, 刘凯. 生成模型在蛋白质序列设计中的应用[J]. 应用化学, 2022, 39(1): 3-17.
WU Qing-Lin, REN Yu-Bin, ZHAI Xiao-Wei, CHEN Dong, LIU Kai. Protein Sequence Design Using Generative Models[J]. Chinese Journal of Applied Chemistry, 2022, 39(1): 3-17.

图1 蛋白质设计方法A.基于模板的配体结合蛋白的设计[31];B.靶蛋白结合物的从头设计[35]
Fig.1 Protein design methodsA. Template-based design of ligand-binding protein[31]; B. De novo design of a binder to a target protein[35]

图2 针对固定骨架的蛋白序列设计A.一般步骤[38];B.基于能量最低和概率最大的两种评价方法[36]
Fig. 2 Design sequence for a given backboneA. General steps[38]; B. Two evaluation methods based on energy minimization or probability maximization[36]
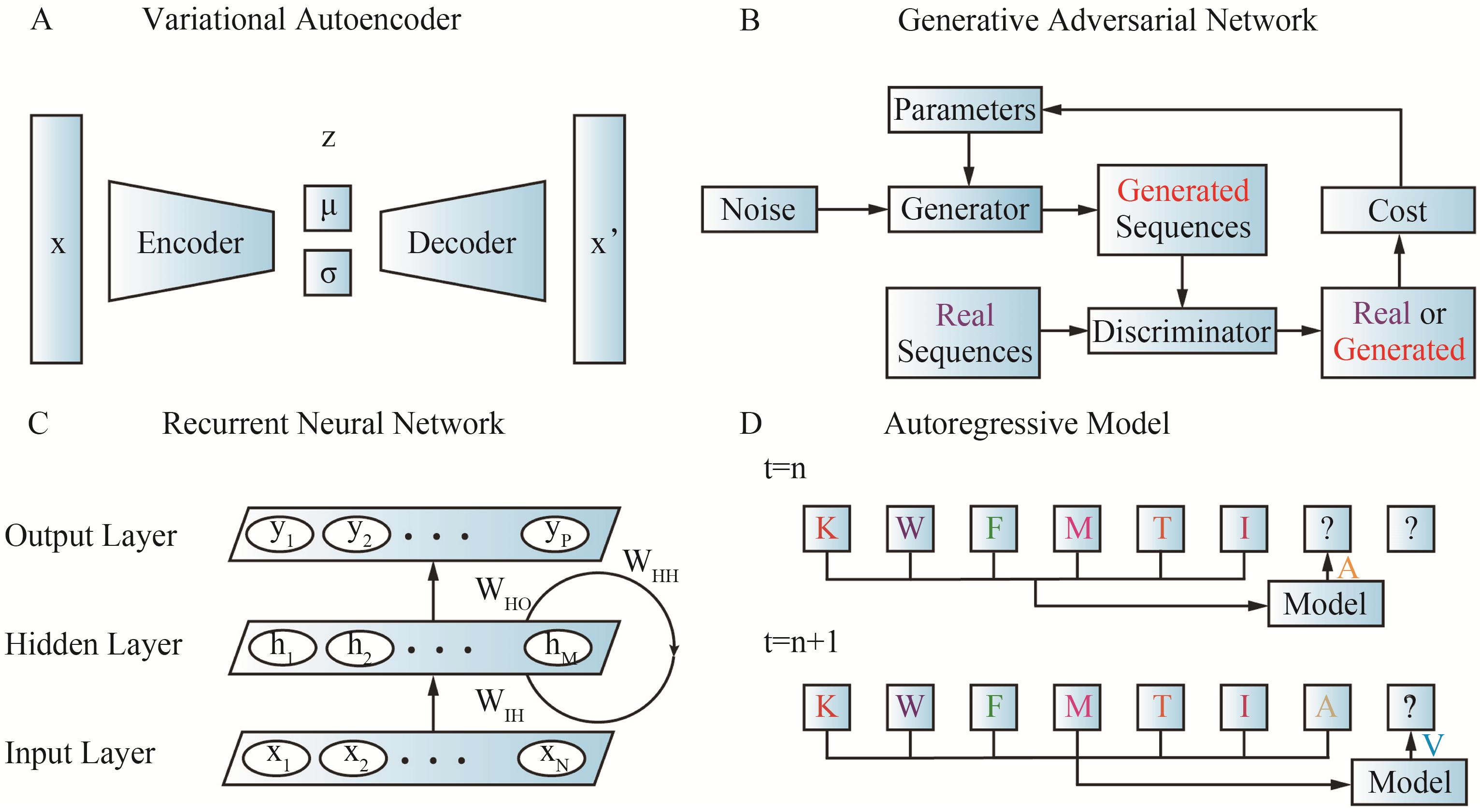
图3 蛋白质序列设计的四种常用生成模型(A)变分自编码器(VAE)[24];(B)生成对抗网络(GAN)[24];(C)循环神经网络(RNN)[48];(D)自回归模型(ARM)[24]
Fig.3 Four commonly used generative models in protein sequence design(A) Variational autoencoder (VAE)[24]; (B) Generative adversarial network (GAN)[24]; (C) Recurrent neural network (RNN)[48]; (D) Autoregressive model (ARM)[24]
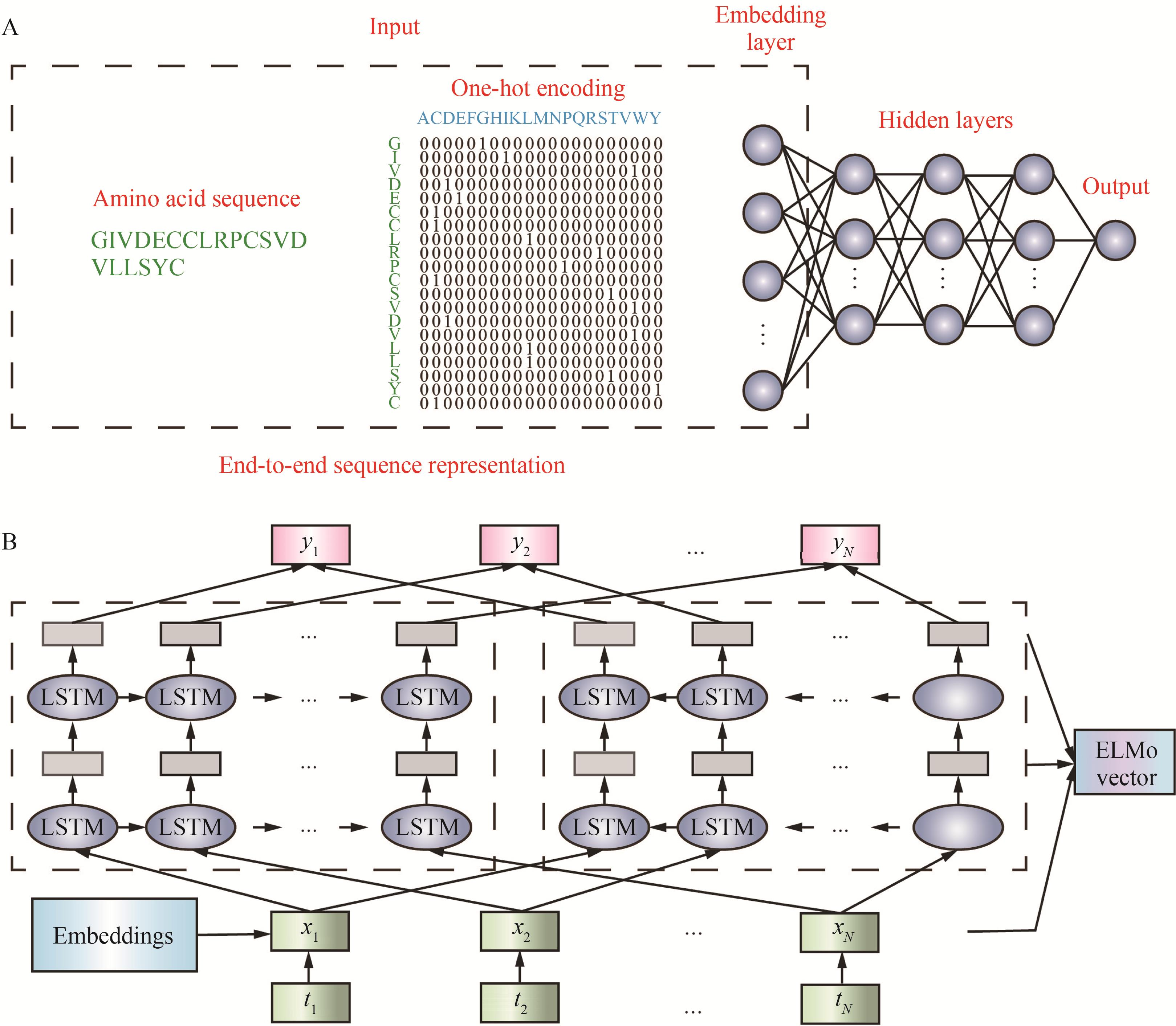
图4 两种常用的蛋白质序列表示方法(A)端到端蛋白序列表示方法[71];(B)ELMo模型表示方法[71]
Fig. 4 Two commonly used methods for protein sequence representation(A) End-to-end sequence representation[71]; (B) Sequence representation by ELMo model[71]
表示方法 Representation methods | 关键机制 Key mechanisms | 优点 Advantages | 缺点 Disadvantages | 应用 Applications |
|---|---|---|---|---|
端到端学习 End-to-end learning | 独热编码 One-hot encoding | 输入直达输出; 获得氨基酸相似性和差异性 Directly from input to output; capture the physicochemical similarities and differences between amino acids | 需要大量高质量有标签的数据 Need a lot of high quality labeled datas | 蛋白质功能预测、分类等 Protein function prediction, classification, etc. |
迁移学习 Transfer Learning | LSTM(ELMo、BERT) | 利用无标签数据;效果好 Using unlabeled data; good performance | 二级结构、蛋白质功能、 蛋白质演化、相互作用等 Secondary structure, protein function, protein evolution, interaction, etc. | |
上下文无关学习 Non-contextual embedding learning | word2vec等 word2vec et al | 性能不佳 Poor performance | 蛋白质分类、功能识别等 Protein classification, functional recognition, etc. | |
其它学习 Other representation methods | GCN、基于特征提取的方法 GCN, feature extraction based method | 应用局限在特定领域 Applications are limited to specific domains | 蛋白质组、蛋白药物、 蛋白质结构等特定领域 Roteome, protein medicine, protein structure and other specific fields |
表1 四类蛋白质序列表示方法的比较
Table1 Comparison of four types of protein sequence representation
表示方法 Representation methods | 关键机制 Key mechanisms | 优点 Advantages | 缺点 Disadvantages | 应用 Applications |
|---|---|---|---|---|
端到端学习 End-to-end learning | 独热编码 One-hot encoding | 输入直达输出; 获得氨基酸相似性和差异性 Directly from input to output; capture the physicochemical similarities and differences between amino acids | 需要大量高质量有标签的数据 Need a lot of high quality labeled datas | 蛋白质功能预测、分类等 Protein function prediction, classification, etc. |
迁移学习 Transfer Learning | LSTM(ELMo、BERT) | 利用无标签数据;效果好 Using unlabeled data; good performance | 二级结构、蛋白质功能、 蛋白质演化、相互作用等 Secondary structure, protein function, protein evolution, interaction, etc. | |
上下文无关学习 Non-contextual embedding learning | word2vec等 word2vec et al | 性能不佳 Poor performance | 蛋白质分类、功能识别等 Protein classification, functional recognition, etc. | |
其它学习 Other representation methods | GCN、基于特征提取的方法 GCN, feature extraction based method | 应用局限在特定领域 Applications are limited to specific domains | 蛋白质组、蛋白药物、 蛋白质结构等特定领域 Roteome, protein medicine, protein structure and other specific fields |

图6 两种优化蛋白质序列的方法(A)基于反馈机制的生成对抗网络(FBGAN)[83]; (B)强化学习(RL)中智能体和环境的交互[88]
Fig.6 Two methods for optimizing protein sequences(A) Feedback generative adversarial network (FBGAN)[83]; (B) Agent-environment interactions in reinforcement learning (RL)[88]
| 1 | HUANG P S, BOYKEN S E, BAKER D. The coming of age of de novo protein design[J]. Nature, 2016, 537(7620): 320-327. |
| 2 | GAINZA-CIRAUQUI P, CORREIA B E. Computational protein design—the next generation tool to expand synthetic biology applications[J]. Curr Opin Biotechnol, 2018, 52: 145-152. |
| 3 | CHEVALIER A, SILVA D A, ROCKLIN G J, et al. Massively parallel de novo protein design for targeted therapeutics[J]. Nature, 2017, 550(7674): 74-79. |
| 4 | KAMERZELL T J, MIDDAUGH C R. Prediction machines: applied machine learning for therapeutic protein design and development[J]. J Pharm Sci, 2021, 110(2): 665-681. |
| 5 | SILVA D A, YU S, ULGE U Y, et al. De novo design of potent and selective mimics of IL-2 and IL-15[J]. Nature, 2019, 565(7738): 186-191. |
| 6 | LI J, LI B, SUN J, et al. Engineered near-infrared fluorescent protein assemblies for robust bioimaging and therapeutic applications[J]. Adv Mater, 2020, 32(17): 2000964. |
| 7 | SUN J, LI B, WANG F, et al. Proteinaceous fibers with outstanding mechanical properties manipulated by supramolecular interactions[J]. CCS Chem, 2021, 3(6): 1669-1677. |
| 8 | XIAO L, WANG Z, SUN Y, et al. An artificial phase-transitional underwater bioglue with robust and switchable adhesion performance[J]. Angew Chem Int Ed, 2021, 60(21): 12082-12089. |
| 9 | ANAND-ACHIM N, EGUCHI R R, MATHEWS I I, et al. Protein sequence design with a learned potential[J]. bioRxiv, 2021,DOI: 10.1101/2020.01.06.895466 |
| 10 | LI Y, LI J, SUN J, et al. Bioinspired and mechanically strong fibers based on engineered non-spider chimeric proteins[J]. Angew Chem Int Ed, 2020, 132(21): 8225-8229. |
| 11 | KUHLMAN B, BRADLEY P. Advances in protein structure prediction and design[J]. Nat Rev Mol Cell Biol, 2019, 20(11): 681-697. |
| 12 | CHEN I M A, MARKOWITZ V M, CHU K, et al. IMG/M: integrated genome and metagenome comparative data analysis system[J]. Nucleic Acids Res, 2017, 45(D1): D507-D516. |
| 13 | WESTBROOK J D, BURLEY S K. How structural biologists and the protein data bank contributed to recent FDA new drug approvals[J]. Structure, 2019, 27(2): 211-217. |
| 14 | LECUN Y, BENGIO Y, HINTON G. Deep learning[J]. Nature, 2015, 521(7553): 436-444. |
| 15 | TUNYASUVUNAKOOL K, ADLER J, WU Z, et al. Highly accurate protein structure prediction for the human proteome[J]. Nature, 2021, 596(7873): 590-596. |
| 16 | YANG J Y, ANISHCHENKO I, PARK H, et al. Improved protein structure prediction using predicted interresidue orientations[J]. Proc Natl Acad Sci USA, 2020, 117(3): 1496-1503. |
| 17 | STROKACH A, BECERRA D, CORBI-VERGE C, et al. Fast and flexible protein design using deep graph neural networks[J]. Cell Syst, 2020, 11(4): 402-4114. |
| 18 | WEI K Y, MOSCHIDI D, BICK M J, et al. Computational design of closely related proteins that adopt two well-defined but structurally divergent folds[J]. Proc Natl Acad Sci USA, 2020, 117(13): 7208-7215. |
| 19 | BAEK M, DIMAIO F, ANISHCHENKO I, et al. Accurate prediction of protein structures and interactions using a three-track neural network[J]. Science, 2021, 373(6557): 871-876. |
| 20 | CUNNINGHAM J M, KOYTIGER G, SORGER P K, et al. Biophysical prediction of protein-peptide interactions and signaling networks using machine learning[J]. Nat Methods, 2020, 17(2): 175-183. |
| 21 | HOPF T A, INGRAHAM J B, POELWIJK F J, et al. Mutation effects predicted from sequence co-variation[J]. Nat Biotechnol, 2017, 35(2): 128-135. |
| 22 | RIESSELMAN A J, INGRAHAM J B, MARKS D S. Deep generative models of genetic variation capture the effects of mutations[J]. Nat Methods, 2018, 15(10): 816-822. |
| 23 | WANG D D, LE O Y, XIE H R, et al. Predicting the impacts of mutations on protein-ligand binding affinity based on molecular dynamics simulations and machine learning methods[J]. Comput Struct Biotechnol J, 2020, 18: 439-454. |
| 24 | WU Z, JOHNSTON K E, ARNOLD F H, et al. Protein sequence design with deep generative models[J]. Curr Opin Chem Biol, 2021, 65: 18-27. |
| 25 | SINAI S, KELSIC E, CHURCH G M, et al. Variational auto⁃encoding of protein sequences[J]. arXiv e⁃prints, 2017: arXiv:. |
| 26 | Bitard⁃Feildel T. Navigating the amino acid sequence space between functional proteins using a deep learning framework[J]. bioRxiv, 2021: 2020.11. 09.375311. DOI: https://doi.org/10.1101/2020.11.09.375311 |
| 27 | REPECKA D, JAUNISKIS V, KARPUS L, et al. Expanding functional protein sequence spaces using generative adversarial networks[J]. Nat Mach Intell, 2021, 3(4): 324-333. |
| 28 | MÜLLER A T, HISS J A, SCHNEIDER G. Recurrent neural network model for constructive peptide design[J]. J Chem Inf Model, 2018, 58(2): 472-479. |
| 29 | SHIN J E, RIESSELMAN A J, KOLLASCH A W, et al. Protein design and variant prediction using autoregressive generative models[J]. Nat Commun, 2021, 12(1): 1-11. |
| 30 | COLUZZA I. Computational protein design: a review[J]. J Phys: Condens Matter, 2017, 29(14): 143001. |
| 31 | TINBERG C E, KHARE S D, DOU J Y, et al. Computational design of ligand-binding proteins with high affinity and selectivity[J]. Nature, 2013, 501(7466): 212-216. |
| 32 | KOEPNICK B, FLATTEN J, HUSAIN T, et al. De novo protein design by citizen scientists[J]. Nature, 2019, 570(7761): 390-394. |
| 33 | YANG C, SESTERHENN F, BONET J, et al. Bottom-up de novo design of functional proteins with complex structural features[J]. Nat Chem Biol, 2021, 17(4): 492-500. |
| 34 | DAWSON W M, RHYS G G, WOOLFSON D N. Towards functional de novo designed proteins[J]. Curr Opin Chem Biol, 2019, 52: 102-111. |
| 35 | PAN X, KORTEMME T. Recent advances in de novo protein design: principles, methods, and applications[J]. J Biol Chem, 2021, 296: 100558. |
| 36 | NORN C, WICKY B I M, JUERGENS D, et al. Protein sequence design by conformational landscape optimization[J]. Proc Natl Acad Sci USA, 2021, 118(11): e2017228118. |
| 37 | SANDHYA S, MUDGAL R, KUMAR G, et al. Protein sequence design and its applications[J]. Curr Opin Struct Biol, 2016, 37: 71-80. |
| 38 | LIU H Y, CHEN Q. Computational protein design for given backbone: recent progresses in general method-related aspects[J]. Curr Opin Struct Biol, 2016, 39: 89-95. |
| 39 | MAGUIRE J B, HADDOX H K, STRICKLAND D, et al. Perturbing the energy landscape for improved packing during computational protein design[J]. Proteins Struct Funct Bioinf, 2021, 89(4): 436-449. |
| 40 | XIONG P, HU X H, HUANG B, et al. Increasing the efficiency and accuracy of the ABACUS protein sequence design method[J]. Bioinformatics, 2020, 36(1): 136-144. |
| 41 | KINGMA D P, WELLING M. Auto⁃encoding variational bayes[J]. arXiv e⁃prints, 2013: arXiv:. |
| 42 | HAWKINS-HOOKER A, DEPARDIEU F, BAUR S, et al. Generating functional protein variants with variational autoencoders[J]. PLoS Comput Biol, 2021, 17(2): e1008736. |
| 43 | GOODFELLOW I, POUGET-ABADIE J, MIRZA M, et al. Generative adversarial nets[J]. Proc Adv Neural Inf Process Syst, 2014, 27. |
| 44 | MIRZA M, OSINDERO S. Conditional generative adversarial nets[J]. arXiv preprint arXiv, 2014, 1411.1784. |
| 45 | ARJOVSKY M, BOTTOU L. Towards principled methods for training generative adversarial networks[J]. arXiv e⁃prints, 2017: arXiv:. |
| 46 | ARJOVSKY M, CHINTALA S, BOTTOU L. Wasserstein generative adversarial networks[C]//International conference on machine learning. PMLR, 2017: 214-223. |
| 47 | YU L, ZHANG W, WANG J, et al. Seqgan: sequence generative adversarial nets with policy gradient[C]//Proceedings of the AAAI conference on artificial intelligence. 2017. |
| 48 | SALEHINEJAD H, SANKAR S, BARFETT J, et al. Recent advances in recurrent neural networks[J]. arXiv e⁃prints, 2017: arXiv:. |
| 49 | TRAN K, BISAZZA A, MONZ C. Recurrent memory networks for language modeling[J]. arXiv e⁃prints, 2016: arXiv:. |
| 50 | MIROWSKI P, VLACHOS A. Dependency recurrent neural language models for sentence completion[J]. arXiv e⁃prints, 2015: arXiv:. |
| 51 | LE Q V, JAITLY N, HINTON G E. A simple way to initialize recurrent networks of rectified linear units[J]. arXiv e⁃prints, 2015: arXiv:. |
| 52 | SHERSTINSKY A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network[J]. Phys D, 2020, 404: 132306. |
| 53 | JÓZEFOWICZ R, ZAREMBA W, SUTSKEVER I. An empirical exploration of recurrent network architectures[C]//International conference on machine learning. PMLR, 2015: 2342-2350. |
| 54 | GRISONI F, SCHNEIDER G. De novo molecular design with generative long short-term memory[J]. Chimia, 2019, 73(12): 1006-1011. |
| 55 | SCHNEIDER P, WALTERS W P, PLOWRIGHT A T, et al. Rethinking drug design in the artificial intelligence era[J]. Nat Rev Drug Discovery, 2020, 19(5): 353-364. |
| 56 | XU M Y, RAN T, CHEN H M. De novo molecule design through the molecular generative model conditioned by 3D information of protein binding sites[J]. J Chem Inf Model, 2021, 61(7): 3240-3254. |
| 57 | CHEN X, MISHRA N, ROHANINEJAD M, et al. Pixelsnail: an improved autoregressive generative model[C]//International Conference on Machine Learning. PMLR, 2018: 864-872. |
| 58 | RIESSELMAN A, SHIN J⁃E, KOLLASCH A, et al. Accelerating protein design using autoregressive generative models[J]. bioRxiv, 2021: 757252. DOI: https://doi.org/10.1101/757252. |
| 59 | BENGIO Y, COURVILLE A, VINCENT P. Representation learning: a review and new perspectives[J]. IEEE Trans Pattern Anal Mach Intell, 2013, 35(8): 1798-1828. |
| 60 | JURTZ V I, JOHANSEN A R, NIELSEN M, et al. An introduction to deep learning on biological sequence data: examples and solutions[J]. Bioinformatics, 2017, 33(22): 3685-3690. |
| 61 | LIU X R, HONG Z Y, LIU J, et al. Computational methods for identifying the critical nodes in biological networks[J]. Briefings Bioinf, 2020, 21(2): 486-497. |
| 62 | HEFFERNAN R, YANG Y D, PALIWAL K, et al. Capturing non-local interactions by long short-term memory bidirectional recurrent neural networks for improving prediction of protein secondary structure, backbone angles, contact numbers and solvent accessibility[J]. Bioinformatics, 2017, 33(18): 2842-2849. |
| 63 | LIU Z, JIN J, CUI Y, et al. DeepSeqPanII: an interpretable recurrent neural network model with attention mechanism for peptide-HLA class II binding prediction[J]. IEEE/ACM Trans Comput Biol Bioinf 2021. DOI:10.1109/TCBB.2021.3074927 |
| 64 | ALLEY E C, KHIMULYA G, BISWAS S, et al. Unified rational protein engineering with sequence-based deep representation learning[J]. Nat Methods, 2019, 16(12): 1315-1322. |
| 65 | ELNAGGAR A, HEINZINGER M, DALLAGO C, et al. ProtTrans: towards cracking the language of life's code through self⁃supervised deep learning and high performance computing[J]. arXiv e⁃prints, 2020: arXiv:. |
| 66 | RIVES A, MEIER J, SERCU T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences[J]. Proc Natl Acad Sci USA, 2021, 118(15): e2016239118. |
| 67 | RAO R, LIU J, VERKUIL R, et al. Msa transformer[J]. bioRxiv, 2021: 2021.02.12.430858. DOI: https://doi.org/10.1101/2021.02.12.430858. |
| 68 | SERCU T, VERKUIL R, MEIER J, et al. Neural Potts Model[J]. bioRxiv, 2021: 2021.04.08.439084. DOI: https://doi.org/10.1101/2021.04.08.439084. |
| 69 | ELABD H, BROMBERG Y, HOARFROST A, et al. Amino acid encoding for deep learning applications[J]. BMC Bioinf, 2020, 21(1): 1-14. |
| 70 | PETERS M E, NEUMANN M, IYYER M, et al. Deep contextualized word representations[J]. arXiv e⁃prints, 2020: arXiv:. |
| 71 | CUI F F, ZHANG Z L, ZOU Q. Sequence representation approaches for sequence-based protein prediction tasks that use deep learning[J]. Briefings Funct Genomics, 2021, 20(1): 61-73. |
| 72 | HEINZINGER M, ELNAGGAR A, WANG Y, et al. Modeling aspects of the language of life through transfer-learning protein sequences[J]. BMC Bioinf, 2019, 20(1): 723. |
| 73 | CHAROENKWAN P, NANTASENAMAT C, HASAN M M, et al. BERT4Bitter: a bidirectional encoder representations from transformers (BERT)-based model for improving the prediction of bitter peptides[J]. Bioinformatics, 2021, 37(17): 2556-2562. |
| 74 | GREENER J G, MOFFAT L, JONES D T. Design of metalloproteins and novel protein folds using variational autoencoders[J]. Sci Rep, 2018, 8(1): 16189. |
| 75 | VASWANI A, SHAZEER N, PARMAR N, et al. Attention is all you need[C]//Advances in neural information processing systems. 2017: 5998-6008. |
| 76 | SURANA S, ARORA P, SINGH D, et al. PandoraGAN: Generating antiviral peptides using Generative Adversarial Network[J]. bioRxiv, 2021: 2021.02.15.431193. DOI: https://doi.org/10.1101/2021.02.15.431193 |
| 77 | YANG R, WU F, ZHANG C, et al. iEnhancer-GAN: a deep learning framework in combination with word embedding and sequence generativeadversarial net to identify enhancers and their strength[J]. Int J Mol Sci, 2021, 22(7): 3589. |
| 78 | CAPECCHI A, CAI X G, PERSONNE H, et al. Machine learning designs non-hemolytic antimicrobial peptides[J]. Chem Sci, 2021, 12(26): 9221-9232. |
| 79 | MAKHZANI A, SHLENS J, JAITLY N, et al. Adversarial autoencoders[J]. arXiv e⁃prints, 2015: arXiv:. |
| 80 | RUSS W P, FIGLIUZZI M, STOCKER C, et al. An evolution-based model for designing chorismate mutase enzymes[J]. Science, 2020, 369(6502): 440-445. |
| 81 | BROOKES D, PARK H, LISTGARTEN J. Conditioning by adaptive sampling for robust design[C]//International conference on machine learning. PMLR, 2019: 773-782. |
| 82 | ANGERMUELLER C, DOHAN D, BELANGER D, et al. Model-based reinforcement learning for biological sequence design[C]//International conference on learning representations. 2019. |
| 83 | GUPTA A, ZOU J. Feedback GAN for DNA optimizes protein functions[J]. Nat Mach Intell, 2019, 1(2): 105-111. |
| 84 | AMIMEUR T, SHAVER J M, KETCHEM R R, et al. Designing feature-controlled humanoid antibody discovery libraries using generative adversarial networks[J]. bioRxiv, 2020. |
| 85 | WAN C, JONES D T. Protein function prediction is improved by creating synthetic feature samples with generative adversarial networks[J]. Nat Mach Intell, 2020, 2(9): 540-550. |
| 86 | LINDER J, BOGARD N, ROSENBERG A B, et al. A generative neural network for maximizing fitness and diversity of synthetic DNA and protein sequences[J]. Cell Syst, 2020, 11(1): 49-62.e16. |
| 87 | RYPEŚĆ G, LEPAK Ł, WAWRZYŃSKI P. Reinforcement learning for on-line sequence transformation[J]. arXiv preprint arXiv, 2105.14097, 2021. |
| 88 | FRANÇOIS-LAVET V, HENDERSON P, ISLAM R, et al. An introduction to deep reinforcement learning[J]. Found Trends Mach Learn, 2018, 11(3-4): 219-354. |
| 89 | POPOVA M, ISAYEV O, TROPSHA A. Deep reinforcement learning for de novo drug design[J]. Sci Adv, 2018, 4(7): eaap7885. |
| 90 | ZHOU Z P, KEARNES S, LI L, et al. Optimization of molecules via deep reinforcement learning[J]. Sci Rep, 2019, 9(1): 10752. |
| 91 | JEON W, KIM D. Autonomous molecule generation using reinforcement learning and docking to develop potential novel inhibitors[J]. Sci Rep, 2020, 10(1): 1-11. |
| 92 | YU H, SCHRECK J S, MA W. Deep reinforcement learning for protein folding in the hydrophobic⁃polar model with pull moves[C]//Third Workshop on Machine Learning and the Physical Sciences, Vancouver, Canada, 2020. |
| [1] | 赵常利, 秦明高, 窦晓秋, 冯传良. 纳米颗粒增强的手性超分子水凝胶成骨性能[J]. 应用化学, 2022, 39(1): 177-187. |
| [2] | 槐梦娇, 刘陶雪婷, 姜忠义. 水通道蛋白启发下的人工水通道研究进展[J]. 应用化学, 2022, 39(1): 99-109. |
| [3] | 江富祥, 刘巧珍, 王果, 陈河如. 由1,5-二羟基多羟基烃合成哌啶衍生物[J]. 应用化学, 2013, 30(07): 769-775. |
| [4] | 李彗晶, 王海水. 硼酸对赖氨酸盐酸盐在半胱氨酸自组装膜上晶体生长的影响[J]. 应用化学, 2012, 29(09): 1041-1045. |
| [5] | 刘倩, 卢俊瑞, 辛春伟, 鲍秀荣, 刘玉清, 朱姗姗, 邹敏. N-(5-氯-2-羟苯基)氨基酸酯的合成、表征及抑菌活性[J]. 应用化学, 2010, 27(09): 1012-1016. |
| [6] | 蔡冬清, 吴正岩, 吴林, 吴跃进, 朱剑豪, 余增亮. 低能量离子束轰击对碳纳米管中杂质的去除[J]. 应用化学, 2010, 27(08): 987-989. |
| [7] | 冯乙巳, 董文杰, 张博, 汤琳, 谭问非, 许华建. 天然海洋环酯肽Stereocalpin A中间体的合成[J]. 应用化学, 2010, 27(02): 240-242. |
| [8] | 阮湘元,曾绍汉, 蔡明招, 徐经伟. GSH-Cd(Ⅱ)、Cu(Ⅱ)配合物在硅片表面形貌的AFM 分析[J]. 应用化学, 2009, 26(12): 1498-1500. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
