
Chinese Journal of Applied Chemistry ›› 2022, Vol. 39 ›› Issue (10): 1475-1487.DOI: 10.19894/j.issn.1000-0518.220032
• Review • Next Articles
Advances in Receptor‑ligand Interactions on Cell Surface Based on Framework Nucleic Acids
Lin-Jie GUO1,2,3, Hong-Zhen PENG2, Jiang LI1,2,3, Li-Hua WANG1,2,3, Ying ZHU1,2,3( )
)
- 1.Division of Physical Biology,CAS Key Laboratory of Interfacial Physics and Technology,Shanghai Institute of Applied Physics,Chinese Academy of Sciences,Shanghai 201800,China
2.The Interdisciplinary Research Center,Shanghai Advanced Research Institute,Chinese Academy of Sciences,Shanghai 201210,China
3.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2022-02-11Accepted:2022-04-14Published:2022-10-01Online:2022-10-05 -
Contact:Ying ZHU -
About author:zhuying@zjlab.org.cn
-
Supported by:the National Natural Science Foundation of China(82050005);the Youth Innovation Promotion Association of CAS(2016236);the Shanghai Municipal Science and Technology Commission(19142202400)
CLC Number:
Cite this article
Lin-Jie GUO, Hong-Zhen PENG, Jiang LI, Li-Hua WANG, Ying ZHU. Advances in Receptor‑ligand Interactions on Cell Surface Based on Framework Nucleic Acids[J]. Chinese Journal of Applied Chemistry, 2022, 39(10): 1475-1487.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: http://yyhx.ciac.jl.cn/EN/10.19894/j.issn.1000-0518.220032

Fig.1 Non-covalent and covalent conjugation between DNA and ligands(a) The cocaine-esterase CocE non-covalently linked to the specified site of DNA origami through streptavidin and biotin interactions[19]; (b) EGFP protein with Histag non-covalently linked to NTA-DNA in the presence of Ni2+[22]; (c) SPDP, SMCC, Snap-tag and Halo-tag mediated covalent conjugation[16]
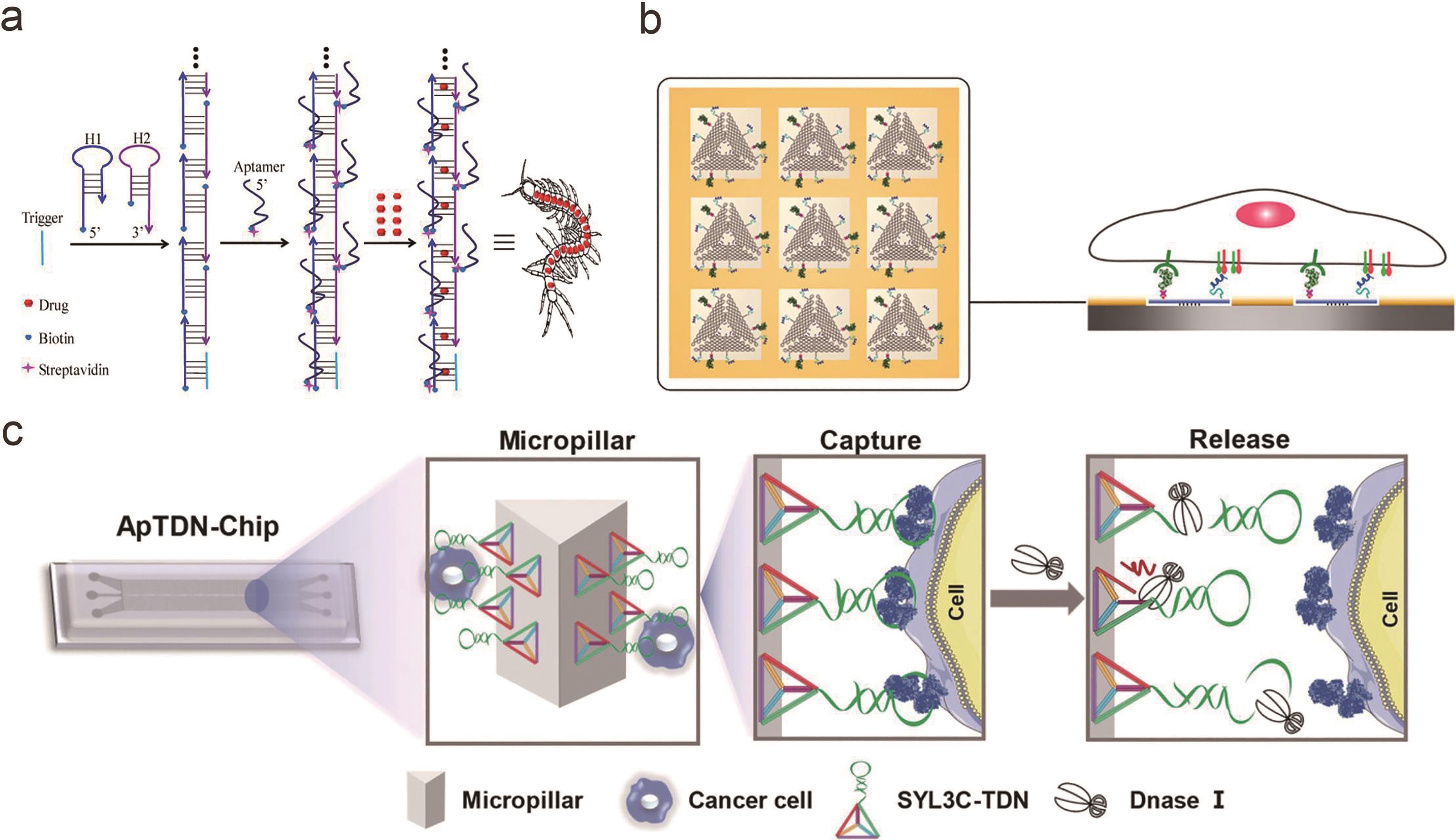
Fig.2 Multivalent interactions between receptor and ligand based on framework nucleic acids(a) Multivalent aptamers conjugated to self-assembled DNA “nanocentipede”[20]; (b) EGF and A20FMDV2 peptides modified on triangular DNA origami for the study of receptor ligand interactions in cancer cell proliferation[42]; (c) DNA tetrahedron with multivalent aptamers on the microfluidic chip interface for circulating tumor cells capture and release[49]

Fig.3 Distance-dependent interactions between receptor and ligand based on framework nucleic acids(a) Design of artificial antigen epitopes with different distances on DNA triangular origami and single-molecule imaging of antigen-antibody interactions under high-speed atomic force microscopy[61]; (b) eOD-GT8 antigens attached to icosahedral and six-helix DNA origami surfaces in varying numbers and intervals[62]; (c) Atomic force microscopy imaging of caspase-9 monomer at different distances in two vertical directions on square DNA origami[63]
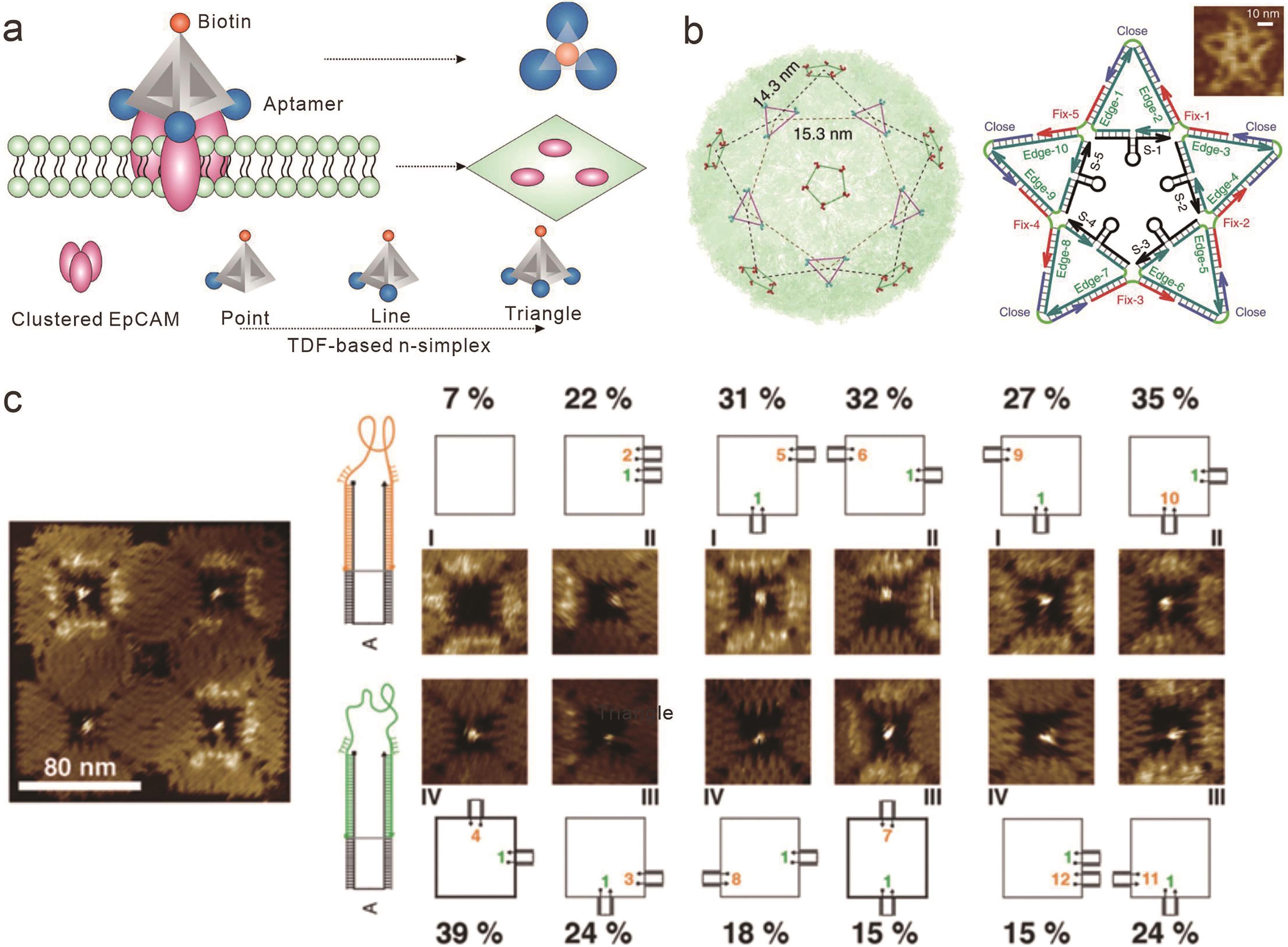
Fig.4 Topologically conformation dependent interactions between receptor and ligand based on framework nucleic acids(a) Topological rearrangement of aptamers based on DNA tetrahedrons and their binding to cell membrane receptors[67]; (b) Star-shaped DNA structure carrying 10 ED3 aptamers based on surface trivalent and pentavalent envelope protein domain Ⅲ (ED3) of dengue virus[69]; (c) Comparison of conformation changes of 11 aptamers and their binding rates with ligands in nanocavities of two-dimensional DNA origami[70]
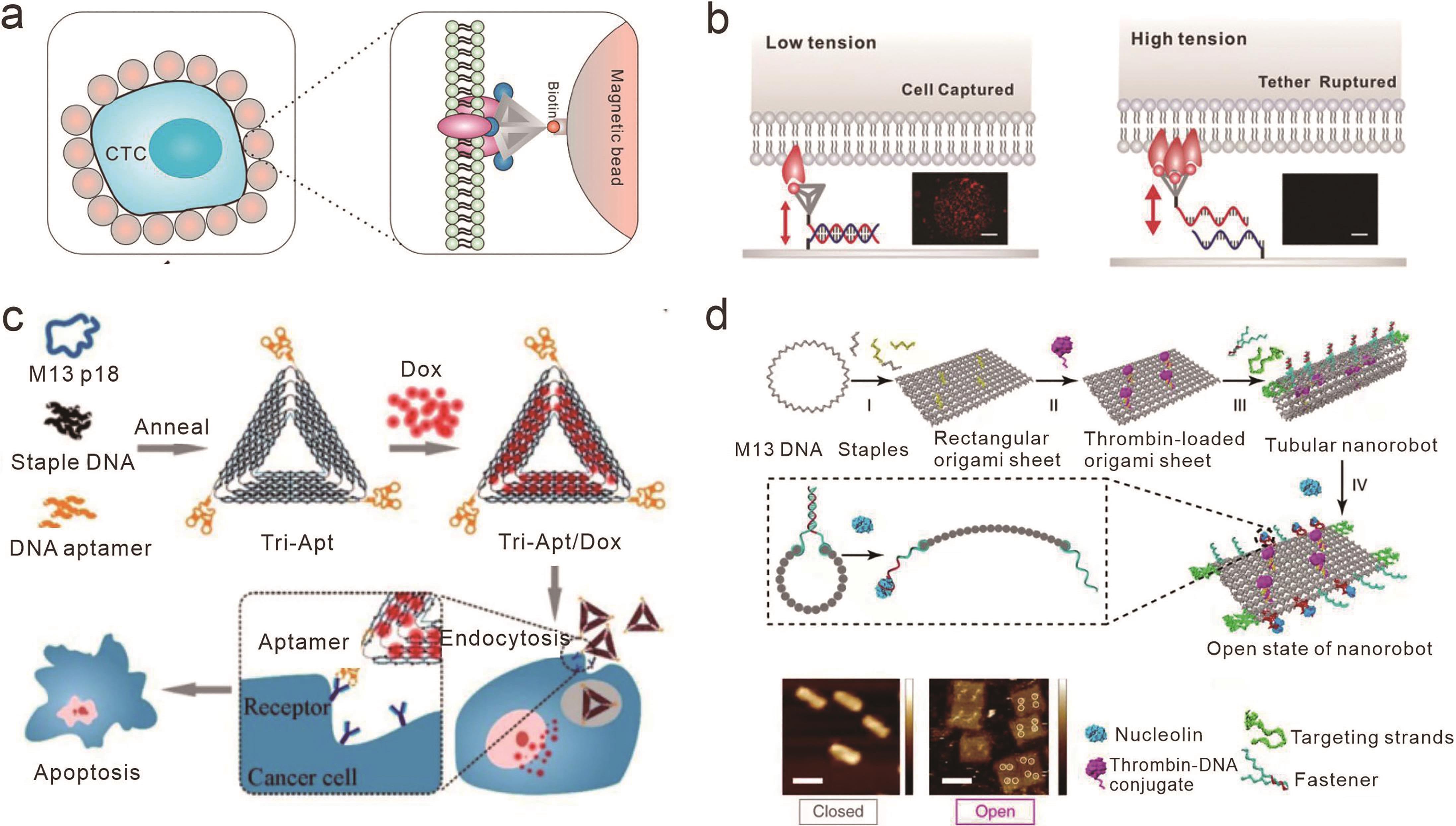
Fig.5 Biological applications of receptor-ligand interactions based on framework nucleic acids(a) Multivalent aptamers based on DNA tetrahedrons for circulating tumor cell capture[67]; (b) Measuring the binding force between multivalent aptamers and cell surface receptors encoded by topological engineering DNA tetrahedrons[68]; (c) Multivalent aptamer-conjugated DNA origami as a targeted delivery vector for the anticancer drug Doxorubicin[75]; (d) Targeted delivery and responsive release of thrombin by DNA nanorobots linked with aptamers[78]
| 1 | KIESSLING L L, GESTWICKI J E.STRONG L E. Synthetic multivalent ligands in the exploration of cell-surface interactions[J]. Curr Opin Chem Biol, 2000, 4: 696-703. |
| 2 | UEKI R, ATSUTA S, UEKI A, et al. Nongenetic reprogramming of the ligand specificity of growth factor receptors by bispecific DNA aptamers[J]. J Am Chem Soc, 2017, 139(19): 6554-6557. |
| 3 | HUPPA J B, AXMANN M, MORTELMAIER M A, et al. Tcr-peptide-MHC interactions in situ show accelerated kinetics and increased affinity[J]. Nature, 2010, 463(7283): 963-967. |
| 4 | TONG G J, HSIAO S C, CARRICO Z M, et al. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles[J]. J Am Chem Soc, 2009, 131: 11174-11178. |
| 5 | YUAN Q, WU Y, WANG J, et al. Targeted bioimaging and photodynamic therapy nanoplatform using an aptamer-guided g-quadruplex DNA carrier and near-infrared light[J]. Angew Chem Int Ed, 2013, 52(52): 13965-13969. |
| 6 | WU Y, SEFAH K, LIU H, et al. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells[J]. Proc Natl Acad Sci U S A, 2010, 107(1): 5-10. |
| 7 | SHENG W, CHEN T, TAN W, et al. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices[J]. ACS Nano, 2013, 7: 7067-7076. |
| 8 | HUANG Y, CHANG H, TAN W. Cancer cell targeting using multiple aptamers conjugated on nanorods[J]. Anal Chem, 2008, 80: 567-572. |
| 9 | GENG Z, WANG L, LIU K, et al. Enhancing anti-PD-1 immunotherapy by nanomicelles self-assembled from multivalent aptamer drug conjugates[J]. Angew Chem Int Ed, 2021, 60(28): 15459-15465. |
| 10 | YANG Y, SUN X, XU J, et al. Circular bispecific aptamer-mediated artificial intercellular recognition for targeted T cell immunotherapy[J]. ACS Nano, 2020, 14(8): 9562-9571. |
| 11 | GE Z, GU H, LI Q, et al. Concept and development of framework nucleic acids[J]. J Am Chem Soc, 2018, 140(51): 17808-17819. |
| 12 | SEEMAN N C, SLEIMAN H F. DNA nanotechnology[J]. Nat Rev Mater, 2017, 3: 17068. |
| 13 | LIU N, LIEDL T. DNA-assembled advanced plasmonic architectures[J]. Chem Rev, 2018, 118(6): 3032-3053. |
| 14 | OUYANG X, WANG M, GUO L, et al. DNA nanoribbon-templated self-assembly of ultrasmall fluorescent copper nanoclusters with enhanced luminescence[J]. Angew Chem Int Ed, 2020, 59(29): 11836-11844. |
| 15 | ZHANG Q, JIANG Q, LI N, et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy[J]. ACS Nano, 2014, 8: 6633-6643. |
| 16 | KONG G, XIONG M, LIU L, et al. DNA origami-based protein networks: from basic construction to emerging applications[J]. Chem Soc Rev, 2021, 50(3): 1846-1873. |
| 17 | KUZUYA A, KIMURA M, NUMAJIRI K, et al. Precisely programmed and robust 2D streptavidin nanoarrays by using periodical nanometer-scale wells embedded in DNA origami assembly[J]. ChemBioChem, 2009, 10(11): 1811-1815. |
| 18 | NUMAJIRI K, KIMURA M, KUZUYA A, et al. Stepwise and reversible nanopatterning of proteins on a DNA origami scaffold[J]. Chem Commun, 2010, 46(28): 5127-5129. |
| 19 | MALLIK L, DHAKAL S, NICHOLS J, et al. Electron microscopic visualization of protein assemblies on flattened DNA origami[J]. ACS Nano, 2015, 9: 7133-7141. |
| 20 | LI W, YANG X, HE L, et al. Self-assembled DNA nanocentipede as multivalent drug carrier for targeted delivery[J]. ACS Appl Mater Interfaces, 2016, 8(39): 25733-25740. |
| 21 | OUYANG X, DE STEFANO M, KRISSANAPRASIT A, et al. Docking of antibodies into the cavities of DNA origami structures[J]. Angew Chem Int Ed, 2017, 56(46): 14423-14427. |
| 22 | SHEN W, ZHONG H, NEFF D, et al. NTA directed protein nanopatterning on DNA origami nanoconstructs[J]. J Am Chem Soc, 2009, 131: 6660-6661. |
| 23 | YAMAZAKI T, HEDDLE J G, KUZUYA A, et al. Orthogonal enzyme arrays on a DNA origami scaffold bearing size-tunable wells[J]. Nanoscale, 2014, 6(15): 9122-9126. |
| 24 | 黄子珂, 刘超, 付强强, 等. 核酸适配体荧光探针在生化分析和生物成像中的研究进展[J]. 应用化学, 2018, 35(1): 28-39. |
| HUANG Z K, LIU C, FU Q Q, et al. Aptamer-based fluorescence probe for bioanalysis and bioimaging[J]. Chinese J Appl Chem, 2018, 35(1): 28-39. | |
| 25 | CHHABRA R, SHARMA J, KE Y, et al. Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures[J]. J Am Chem Soc, 2007(129): 10304-10305. |
| 26 | FU J, LIU M, LIU Y, et al. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures[J]. J Am Chem Soc, 2012, 134(12): 5516-5519. |
| 27 | FISHER P D E, SHEN Q, AKPINAR B, et al. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement[J]. ACS Nano, 2018, 12(2): 1508-1518. |
| 28 | NAKATA E, DINH H, NGO T A, et al. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds[J]. Chem Commun, 2015, 51(6): 1016-1019. |
| 29 | KOSSMANN K J, ZIEGLER C, ANGELIN A, et al. A rationally designed connector for assembly of protein-functionalized DNA nanostructures[J]. ChemBioChem, 2016, 17(12): 1102-1106. |
| 30 | SACCA B, MEYER R, ERKELENZ M, et al. Orthogonal protein decoration of DNA origami[J]. Angew Chem Int Ed, 2010, 49(49): 9378-9383. |
| 31 | ILIUK A B, HU L, TAO W A. Aptamer in bioanalytical applications[J]. Anal Chem, 2011, 83(12): 4440-4452. |
| 32 | TAN W, DONOVAN M J, JIANG J, Aptamers from cell-based selection for bioanalytical applications[J]. Chem Rev, 2013, 113(4): 2842-2862. |
| 33 | MAMMEN M, CHOI S, WHITESIDES G M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors[J]. Angew Chem Int Ed, 1998, 37: 2754-2794. |
| 34 | FASTING C, SCHALLEY C A, WEBER M, et al. Multivalency as a chemical organization and action principle[J]. Angew Chem Int Ed, 2012, 51(42): 10472-10498. |
| 35 | CZAJKOWSKYA D M, SHAO Z. The human IgM pentamer is a mushroom-shaped molecue with a flexural bias[J]. PNAS, 2009, 106: 14960-14965. |
| 36 | HERNANDEZ-LOPEZ R A, YU W, CABRAL K A, et al. T cell circuits that sense antigen density with an ultrasensitive threshold[J]. Science, 2021, 371: 1166-1171. |
| 37 | LIN M, ZHANG J, WAN H, et al. Rationally designed multivalent aptamers targeting cell surface for biomedical applications[J]. ACS Appl Mater Interfaces, 2021, 13(8): 9369-9389. |
| 38 | ZHU G, HU R, ZHAO Z, et al. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications[J]. J Am Chem Soc, 2013, 135(44): 16438-16445. |
| 39 | LIU X, YAN H, LIU Y, et al. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers[J]. Small, 2011, 7(12): 1673-1682. |
| 40 | ROTHEMUND P W. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440(7082): 297-302. |
| 41 | 付衍明, 张钊, 李璨, 等. DNA折纸术研究进展[J]. 应用化学, 2010, 27 (2): 125-131. |
| FU Y M, ZHANG Z, LI C, et al. Recent progress in DNA origami[J]. Chinese J Appl Chem, 2010, 27(2): 125-131. | |
| 42 | HUANG D, PATEL K, PEREZ-GARRIDO S, et al. DNA origami nanoarrays for multivalent investigations of cancer cell spreading with nanoscale spatial resolution and single-molecule control[J]. ACS Nano, 2019, 13(1): 728-736. |
| 43 | HAWKES W, HUANG D, REYNOLDS P, et al. Probing the nanoscale organisation and multivalency of cell surface receptors: DNA origami nanoarrays for cellular studies with single-molecule control[J]. Faraday Discuss, 2019, 219(0): 203-219. |
| 44 | DUTTA P K, ZHANG Y, BLANCHARD A T, et al. Programmable multivalent DNA-origami tension probes for reporting cellular traction forces[J]. Nano Lett, 2018, 18(8): 4803-4811. |
| 45 | GOODMAN R P, BERRY R M, TURBERFIELD A J. The single-step synthesis of a DNA tetrahedron[J]. Chem Commun, 2004(12): 1372-1373. |
| 46 | GOODMAN R P, SCHAAP A T, TARDIN C F, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication[J]. 2005, 310: 1661-1665. |
| 47 | ZHANG T, CUI W, TIAN T, et al. Progress in biomedical applications of tetrahedral framework nucleic acid-based functional systems[J]. ACS Appl Mater Interfaces, 2020, 12(42): 47115-47126. |
| 48 | LI J, PEI H, ZHU B, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides[J]. ACS Nano, 2011, 5: 8783-8789. |
| 49 | ZHANG J, LIN B, WU L, et al. DNA nanolithography enables a highly ordered recognition interface in a microfluidic chip for the efficient capture and release of circulating tumor cells[J]. Angew Chem Int Ed, 2020, 59(33): 14115-14119. |
| 50 | LI J, DAI J, JIANG S, et al. Encoding quantized fluorescence states with fractal DNA frameworks[J]. Nat Commun, 2020, 11(1): 2185 |
| 51 | XIE M, GUO L, XING S, et al. Cell imaging with multi-color DNA framework probes[J]. Chem Commun, 2021, 57(86): 11318-11321. |
| 52 | SIMONS K, TOOMRE D. Lipid rafts and signal transduction[J]. Nat Rev, 2000, 1: 31-41. |
| 53 | GOMES DE CASTRO M A, WILDHAGEN H, SOGRATE-IDRISSI S, et al. Differential organization of tonic and chronic B cell antigen receptors in the plasma membrane[J]. Nat Commun, 2019, 10(1) :820. |
| 54 | DEY S, FAN C, GOTHELF K V, et al. DNA origami[J]. Nat Rev Methods Primers, 2021, 1(1): 13. |
| 55 | RINKER S, KE Y, LIU Y, et al. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding[J]. Nat Nanotechnol, 2008, 3(7): 418-422. |
| 56 | SHAW A, LUNDIN V, PETROVA E, et al. Spatial control of membrane receptor function using ligand nanocalipers[J]. Nat Methods, 2014, 11(8): 841-846. |
| 57 | DIEBOLDER C A, BEURSKENS F J, JONG R N, et al. Complement is activated by IgG hexamers assembled at the cell surface[J]. Science, 2014, 343: 1260-1264. |
| 58 | SONDERMANN P, HUBER R, OOSTHUIZEN V, et al. The 3.2-Å crystal structure of the human IgG fc fragment-fcγriii complex[J]. Nature, 2000, 406: 267-273. |
| 59 | QIAO S, KOBAYASHI K, JOHANSEN F E, et al. Dependence of antibody-mediated presentation of antigen on FcRn[J]. Proc Nat Acad Sci, 2008, 105: 9337-9342. |
| 60 | SHAW A, SANDLIE I, MICHAELSEN T E, et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies[J]. Nat Nanotechnol, 2019, 14: 184-190. |
| 61 | ZHANG P, LIU X, LIU P, et al. Capturing transient antibody conformations with DNA origami epitopes[J]. Nat Commun, 2020, 11(1): 3114. |
| 62 | VENEZIANO R, MOYER T J, STONE M B, et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami[J]. Nat Nanotechnol, 2020, 15(8): 716-723. |
| 63 | ROSIER B, MARKVOORT A J, GUMI AUDENIS B, et al. Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome[J]. Nat Catal, 2020, 3(3): 295-306. |
| 64 | ZHANG K, DENG R, SUN Y, et al. Reversible control of cell membrane receptor function using DNA nano-spring multivalent ligands[J]. Chem Sci, 2017, 8(10): 7098-7105. |
| 65 | HARRISON S C. Looking inside adenovirus[J]. Science, 2010, 329: 1026-1027. |
| 66 | CAI H, MULLER J, DEPOIL D, et al. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering[J]. Nat Nanotechnol, 2018, 13: 610-617. |
| 67 | LI M, DING H, LIN M, et al. DNA framework-programmed cell capture via topology-engineered receptor-ligand interactions[J]. J Am Chem Soc, 2019, 141(47): 18910-18915. |
| 68 | YIN F, LI M, MAO X, et al. DNA framework-based topological cell sorters[J]. Angew Chem Int Ed, 2020, 59(26): 10406-10410. |
| 69 | KWON P S, REN S, KWON S J, et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition[J]. Nat Chem, 2020, 12(1): 26-35. |
| 70 | RAFAT A A, SAGREDO S, THALHAMMER M, et al. Barcoded DNA origami structures for multiplexed optimization and enrichment of DNA-based protein-binding cavities[J]. Nat Chem, 2020, 12: 852-859. |
| 71 | ZHONG L, CAI S, HUANG Y, et al. DNA octahedron-based fluorescence nanoprobe for dual tumor-related mrnas detection and imaging[J]. Anal Chem, 2018, 90(20): 12059-12066. |
| 72 | XUE C, ZHANG S, LI C, et al. Y-shaped backbone-rigidified triangular DNA scaffold-directed stepwise movement of a dnazyme walker for sensitive microrna imaging within living cells[J]. Anal Chem, 2019, 91(24): 15678-15685. |
| 73 | LI Q, ZHAO D, SHAO X, et al. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery[J]. ACS Appl Mater Interfaces, 2017, 9(42): 36695-36701. |
| 74 | SHI S, FU W, LIN S, et al. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier[J]. Nanomedicine, 2019, 21: 102061. |
| 75 | CAO M, SUN Y, XIAO M, et al. Multivalent aptamer-modified DNA origami as drug delivery system for targeted cancer therapy[J]. Chem Res Chinese Univ, 2019, 36(2): 254-260. |
| 76 | GE Z, GUO L, WU G, et al. DNA origami-enabled engineering of ligand-drug conjugates for targeted drug delivery[J]. Small, 2020, 16(16): e1904857. |
| 77 | ZHANG P, FISCHER A, OUYANG Y, et al. Aptamer-modified DNA tetrahedra-gated metal-organic framework nanoparticle carriers for enhanced chemotherapy or photodynamic therapy[J]. Chem Sci, 2021, 12(43): 14473-14483. |
| 78 | LI S, JIANG Q, LIU S, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo[J]. Nat Biotech, 2018, 36: 258-264. |
| [1] | WEI Guobing,KONG Derong,XU Huihui,YIN Zhaojiang,ZHANG Jing,CUI Hanfeng,HONG Nian,YANG Jie,XIONG Wei,LIU Wenming,GUO Qianwen,CHENG Lin,FAN Hao. A New Type of Electrochemiluminescence Sensor for Detection of Mercury Ion in Chinese Medicinal Materials Danshen Based on Host-Guest Interaction [J]. Chinese Journal of Applied Chemistry, 2019, 36(5): 595-602. |
| [2] | ZHU Jiang, GOU Wenxiu, WANG Xiaomin, CHEN Lidong, LENGRubing, JIANG Chunjie. Synthesis of Ionic Coordination Compounds with Naphthalenedisulfonic Acid and Their Molecular Recognition Properties [J]. Chinese Journal of Applied Chemistry, 2017, 34(1): 105-110. |
| [3] | ZHANG Tieli, CHAI Guoping, HE Caixia, WANG Lei. Preparation of p-Hydroxybenzoic Acid-Imprinted Polymer Particulates and Application to Solid Phase Extraction [J]. Chinese Journal of Applied Chemistry, 2016, 33(2): 229-237. |
| [4] | WAGN Susu, ZHANG Yue, LI Hui, XU Miaomiao. Preparation, Characterization and Recognition Behavior of Quercetin-Rutin Bi-template Molecularly Imprinted Polymers [J]. Chinese Journal of Applied Chemistry, 2015, 32(11): 1290-1298. |
| [5] | HOU Changjun1*, ZHANG Yuchan1, HUANG Shun1, LIU Zhen1, SHEN Caihong2, ZHANG Suyi2, HUO Danqun1. Research Progress of Biomolecular Recognition Based on Porphyrins and Porphyrin Derivatives [J]. Chinese Journal of Applied Chemistry, 2011, 28(09): 977-985. |
| [6] | WU Lin-Hua*, TA Hong-Gui, ZHONG Chun-Long. Selective Recognition of Zinc Ion by N'-((2-Hydroxynaphthalen-1-yl)methylene)-2-naphthohydrazide [J]. Chinese Journal of Applied Chemistry, 2010, 27(06): 632-636. |
| [7] | XU Zhi-Feng*, WU Shuang, WU Xiao-Hui, KUANG Dai-Zhi, ZHANG Fu-Xing, WANG Jian-Qiu. Studies on the Preparation, Mechanism of Recognition and Binding Characteristic of Ferrocenecarboxylic Acid Imprinted Polymers [J]. Chinese Journal of Applied Chemistry, 2010, 27(04): 384-389. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||