应用化学 ›› 2023, Vol. 40 ›› Issue (3): 360-373.DOI: 10.19894/j.issn.1000-0518.220229
深度学习在化学信息学中的应用研究进展
刘振邦1, 张硕2, 包宇2( ), 马英明2, 梁蔚淇2, 王伟2, 何颖2, 牛利2
), 马英明2, 梁蔚淇2, 王伟2, 何颖2, 牛利2
- 1.广州大学计算机科学与网络工程学院,广州 510006
2.广州大学化学化工学院分析科学技术研究中心,广州 510006
-
收稿日期:2022-07-01接受日期:2022-11-10出版日期:2023-03-01发布日期:2023-03-27 -
通讯作者:包宇
Progress of Application Research on Cheminformatics in Deep Learning
Zhen-Bang LIU1, Shuo ZHANG2, Yu BAO2( ), Ying-Ming MA2, Wei-Qi LIANG2, Wei WANG2, Ying HE2, Li NIU2
), Ying-Ming MA2, Wei-Qi LIANG2, Wei WANG2, Ying HE2, Li NIU2
- 1.School of Computer Science and Cyber Engineering,Guangzhou University,Guangzhou 510006,China
2.Center for Advanced Analytical Science,School of Chemistry and Chemical Engineering,Guangzhou University,Guangzhou 510006,China
-
Received:2022-07-01Accepted:2022-11-10Published:2023-03-01Online:2023-03-27 -
Contact:Yu BAO -
About author:baoyu@gzhu.edu.cn
摘要:
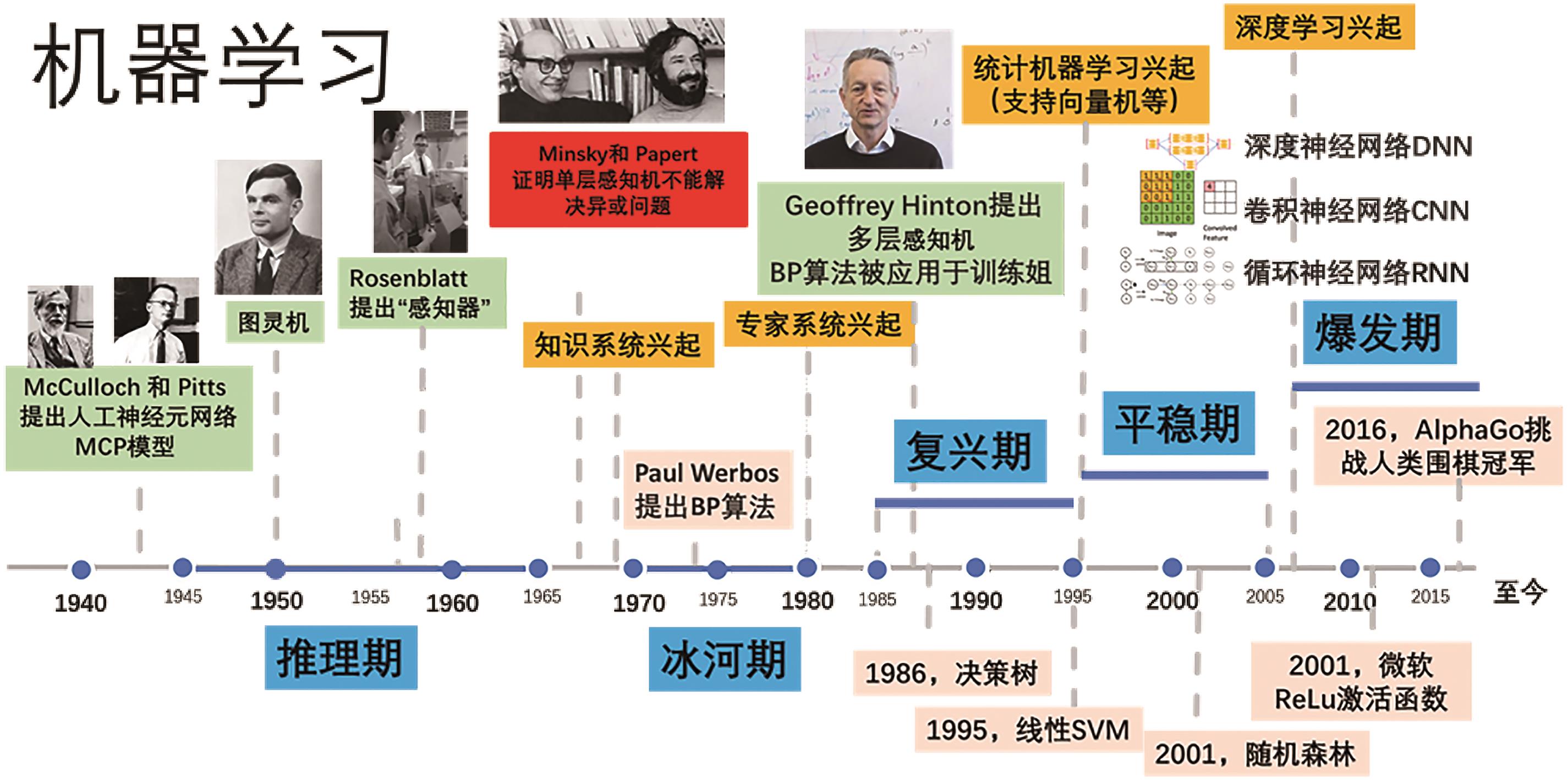
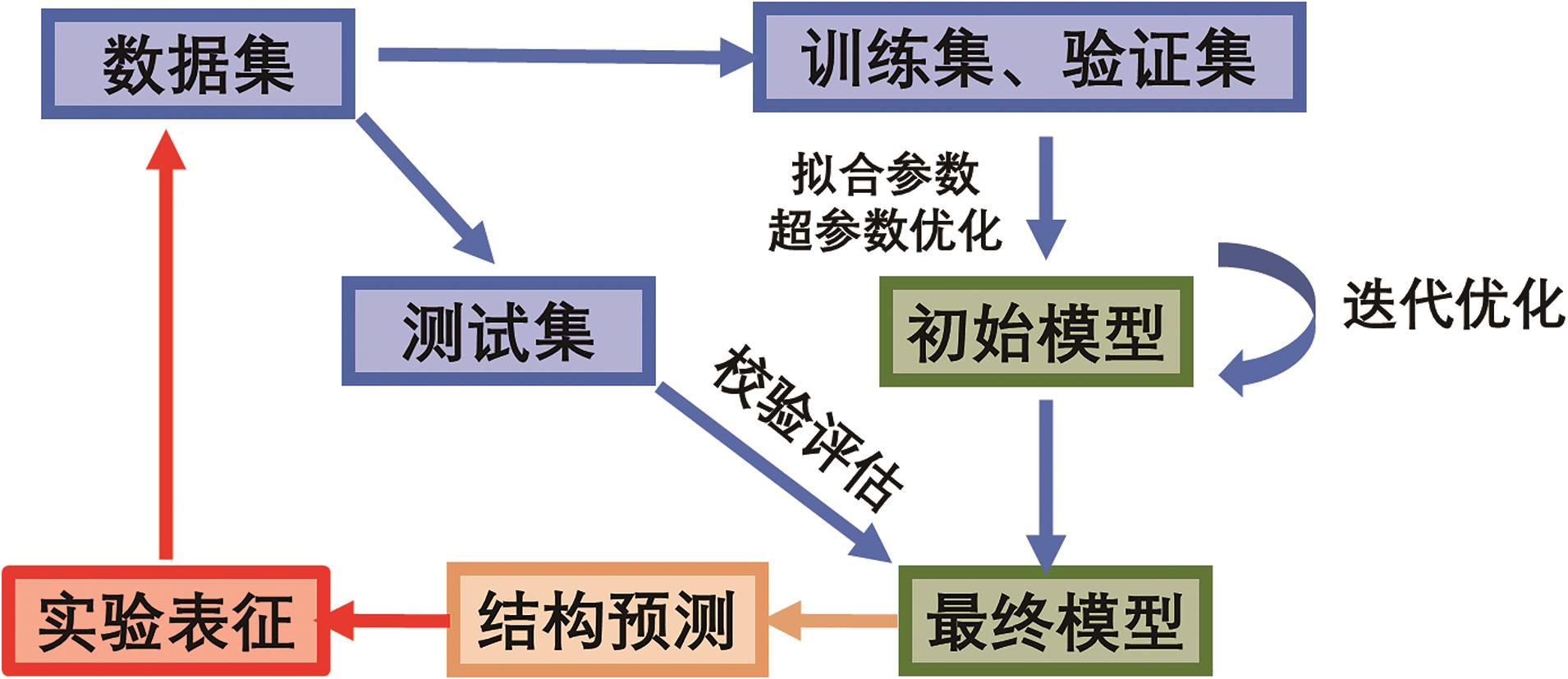
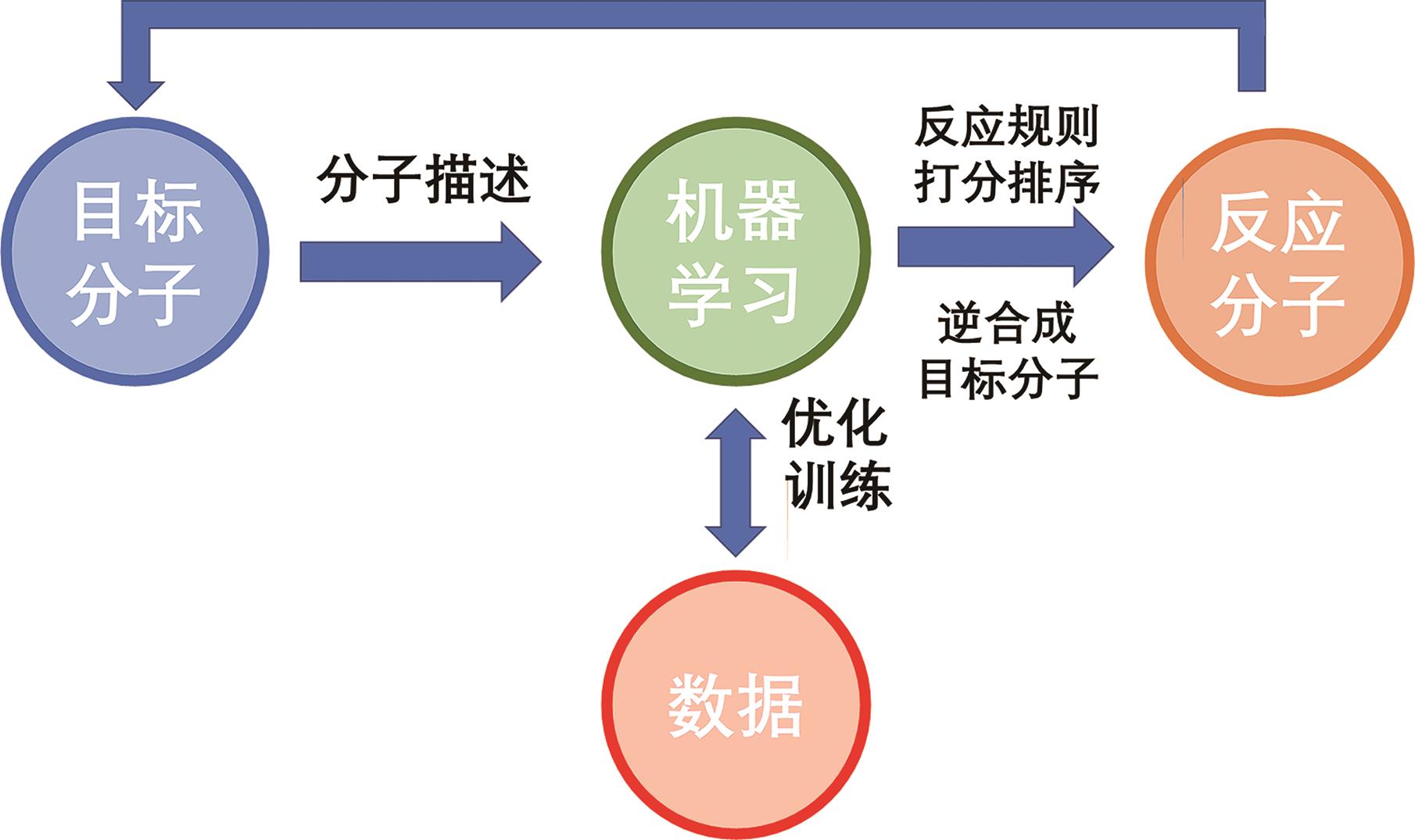
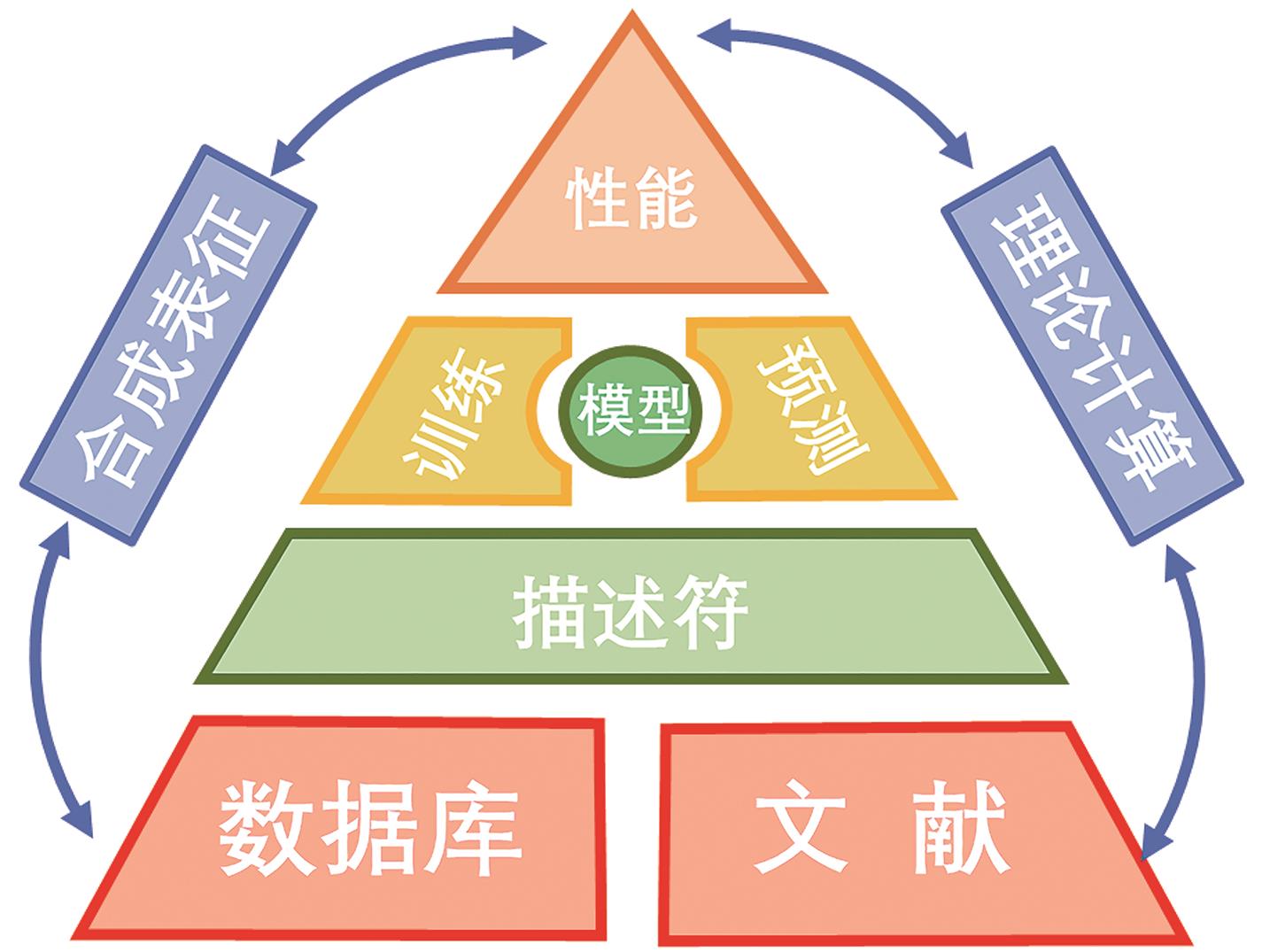
在知识、数据、算法与算力的多重驱动下,深度学习不仅在计算机视觉、自然语言处理等研究领域取得了突破,并随着各学科间的迁移应用于交叉融合,逐渐衍生出多个新兴研究方向。化学信息学作为以应用信息学方法以解决化学问题的学科,深度学习技术凭借其强大的非线性学习能力,通过深度学习模型可以从数据集中对其进行筛选预测,再基于理论计算对结果可行性进行理论验证,最后通过实验表征结果,缩短了实验周期、降低了人力成本、加速了化学信息学智能化。本文简要介绍了深度学习发展历史及主要网络模型架构,介绍了近年来深度学习在在化合物合成路线规划、化合物结构活性与性质及催化剂设计的最新研究和应用现状,并对未来的发展方向进行讨论与展望。
中图分类号:
引用本文
刘振邦, 张硕, 包宇, 马英明, 梁蔚淇, 王伟, 何颖, 牛利. 深度学习在化学信息学中的应用研究进展[J]. 应用化学, 2023, 40(3): 360-373.
Zhen-Bang LIU, Shuo ZHANG, Yu BAO, Ying-Ming MA, Wei-Qi LIANG, Wei WANG, Ying HE, Li NIU. Progress of Application Research on Cheminformatics in Deep Learning[J]. Chinese Journal of Applied Chemistry, 2023, 40(3): 360-373.
| Resear chinterests | Algorithm model | Purpose of study | Datasets | Ref. |
|---|---|---|---|---|
| Structure-activity relationship | DNN, MT-DNN | Prediction of Target structural protein | ChEMBL, A4, B1, PP4, IVINT | [ |
| LSTM-QSAR | Data augmentation with SMILES descriptors | — | [ | |
| LSTM | Generation of model molecular structures | ChEMBL | [ | |
| — | Feature extraction based on molecular fingerprint | The Harvard clean energy project | [ | |
| CNN | Feature extraction of molecular structural formula | Abraham octanol solubility,Delaney small aqueous solubility,Tox21 | [ | |
| Synthesis planning | seq2seq | Machine learning for organic reaction equations | Jin's USPTO | [ |
| seq2seq | Extraction of unsupervised molecular descriptors | — | [ | |
| Expansion policy network, Rollout policy network | Optimization of retrosynthetic routes | Reaxys database | [ | |
| Catalysis chemistry | Transferlearning | Search and design of alloy catalysts | — | [ |
| Decision tree | Prediction of interfacial thermal resistance in nanostructures | — | [ | |
| TPOT | Carbon dioxide reduction in electrocatalysis | GASpy | [ | |
| Gradient boosting regression | Prediction of the activity trends of metal surface | — | [ | |
| Extra tree regression | Prediction of adsorption on alloy surfaces | Surface energies of elemental crystals | [ | |
| CNN | Local structure identification in TEM images | Atomic Simulation Environment | [ | |
| DTNN, AI-Spectroscopist | Prediction of molecular excitation spectra | QM7b,M9 | [ | |
| CNN | Identifying chemical substances using Raman spectroscopy | RRUFF | [ |
表1 深度学习在构效关系、合成路线及催化化学方面的代表性应用
Table 1 Representative applications of deep learning in structure-activity relationships, synthesis planning, and catalysis chemistry
| Resear chinterests | Algorithm model | Purpose of study | Datasets | Ref. |
|---|---|---|---|---|
| Structure-activity relationship | DNN, MT-DNN | Prediction of Target structural protein | ChEMBL, A4, B1, PP4, IVINT | [ |
| LSTM-QSAR | Data augmentation with SMILES descriptors | — | [ | |
| LSTM | Generation of model molecular structures | ChEMBL | [ | |
| — | Feature extraction based on molecular fingerprint | The Harvard clean energy project | [ | |
| CNN | Feature extraction of molecular structural formula | Abraham octanol solubility,Delaney small aqueous solubility,Tox21 | [ | |
| Synthesis planning | seq2seq | Machine learning for organic reaction equations | Jin's USPTO | [ |
| seq2seq | Extraction of unsupervised molecular descriptors | — | [ | |
| Expansion policy network, Rollout policy network | Optimization of retrosynthetic routes | Reaxys database | [ | |
| Catalysis chemistry | Transferlearning | Search and design of alloy catalysts | — | [ |
| Decision tree | Prediction of interfacial thermal resistance in nanostructures | — | [ | |
| TPOT | Carbon dioxide reduction in electrocatalysis | GASpy | [ | |
| Gradient boosting regression | Prediction of the activity trends of metal surface | — | [ | |
| Extra tree regression | Prediction of adsorption on alloy surfaces | Surface energies of elemental crystals | [ | |
| CNN | Local structure identification in TEM images | Atomic Simulation Environment | [ | |
| DTNN, AI-Spectroscopist | Prediction of molecular excitation spectra | QM7b,M9 | [ | |
| CNN | Identifying chemical substances using Raman spectroscopy | RRUFF | [ |
| 1 | SCHMIDHUBER J. Deep learning in neural networks: an overview[J]. Neural Networks, 2015, 61: 85-117. |
| 2 | HINTON G E, OSINDERO S, TEH Y W. A fast learning algorithm for deep belief nets[J]. Neural Comput, 2006, 18(7): 1527-1554. |
| 3 | SILVER D, HUANG A, MADDISON C J, et al. Mastering the game of go with deep neural networks and tree search[J]. Nature, 2016, 529(7587): 484-489. |
| 4 | VINYALS O, BABUSCHKIN I, CZARNECKI W M, et al. Grandmaster level in StarCraft Ⅱ using multi-agent reinforcement learning[J]. Nature, 2019, 575(7782): 350-354. |
| 5 | BAEK M, DIMAIO F, ANISHCHENKO I, et al. Accurate prediction of protein structures and interactions using a three-track neural network[J]. Science, 2021, 373(6557): 871-876. |
| 6 | XIONG P, HU X, HUANG B, et al. Increasing the efficiency and accuracy of the ABACUS protein sequence design method[J]. Bioinformatics, 2020, 36(1): 136-144. |
| 7 | HUANG B, XU Y, HU X, et al. A backbone-centred energy function of neural networks for protein design[J]. Nature, 2022, 602(7897): 523-528. |
| 8 | ROSENBLATT F. Perceptron simulation experiments[J]. Proce IRE, 1960, 48(3): 301-309. |
| 9 | ROSENBLATT F. The perceptron: a probabilistic model for information storage and organization in the brain[J]. Psychol Rev, 1958, 65(6): 386-408. |
| 10 | MINSKY M L, PAPERT S A. Perceptrons: expanded edition[M]. Cambridge, MA, USA: MIT Press, 1988. |
| 11 | PAUL W. Beyond regression: new tools for prediction and analysis in the behavioral sciences[D]. Cambridge, MA, USA: Harvard University, 1974. |
| 12 | KUNIHIKO F. Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position[J]. Biol Cybern, 1980, 36: 193-202. |
| 13 | HOPFIELD J J. Neural networks and physical systems with emergent collective computational abilities[J]. Proc Natl Acad Sci, 1982, 79(8): 2554-2558. |
| 14 | RUMELHART D E, HINTON G E, WILLIAMS R J. Learning representations by back-propagating errors[J]. Nature, 1986, 323(6088): 533-536. |
| 15 | CORTES C, VAPNIK V. Support-vector networks[J]. Mach Learn, 1995, 20(3): 273-297. |
| 16 | HINTON G, DENG L, YU D, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups[J]. IEEE Signal Proc Mag, 2012, 29(6): 82-97. |
| 17 | QUINLAN J R. Induction of decision trees[J]. Mach Learn, 1986, 1(1): 81-106. |
| 18 | SIBSON R. SLINK: an optimally efficient algorithm for the single-link cluster method[J]. Comput J, 1973, 16(1): 30-34. |
| 19 | HO T K. The random subspace method for constructing decision forests[J]. IEEE T Pattern Anal, 1998, 20(8): 832-844. |
| 20 | DOMINGOS P, PAZZANI M. On the optimality of the simple bayesian classifier under zero-one loss[J]. Mach Learn, 1997, 29(2): 103-130. |
| 21 | BENGIO Y. Learning deep architectures for AI[J]. Found Trends Mach Learn, 2009, 2(1): 1-127. |
| 22 | OJALA T, PIETIKAINEN M, MAENPAA T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns[J]. IEEE T Pattern Anal, 2002, 24(7): 971-987. |
| 23 | HÅSTAD J, GOLDMANN M. On the power of small-depth threshold circuits[J]. Comput Complex, 1991, 1(2): 113-129. |
| 24 | BENGIO Y, COURVILLE A, VINCENT P. Representation learning: a review and new perspectives[J]. IEEE T Pattern Anal, 2013, 35(8): 1798-1828. |
| 25 | HORNIK K, STINCHCOMBE M, WHITE H. Multilayer feedforward networks are universal approximators[J]. Neural Networks, 1989, 2(5): 359-366. |
| 26 | ZHANG K, ZUO W, CHEN Y, et al. Beyond a gaussian denoiser: residual learning of deep cnn for image denoising[J]. IEEE T Image Process, 2017, 26(7): 3142-3155. |
| 27 | WERBOS P J. Backpropagation through time: what it does and how to do it[J]. P IEEE, 1990, 78(10): 1550-1560. |
| 28 | WU Z H, PAN S R, CHEN F W, et al. A comprehensive survey on graph neural networks[J]. IEEE Trans Neural Netw Leark, 2021, 32(1): 4-24. |
| 29 | HAZAN E, AGARWAL A, KALE S. Logarithmic regret algorithms for online convex optimization[J]. Mach Learn, 2007, 69(2): 169-192. |
| 30 | GLOROT X, BORDES A, BENGIO Y. Deep sparse rectifier neural networks: proceedings of the fourteenth international conference on artificial intelligence and statistics(PMLR)[C]. Fort Lauderdale, FL, USA, 2011: 315-323. |
| 31 | SRIVASTAVA N, HINTON G, KRIZHEVSKY A, et al. Dropout: a simple way to prevent neural networks from overfitting[J]. J Mach Learn Res, 2014, 15(1): 1929-1958. |
| 32 | HE K, ZHANG X, REN S, et al. Deep residual learning for image recognition: proceedings of the IEEE conference on computer vision and pattern recognition(CVPR)[C]. Las Vegas, NV, USA, 2016: 770-778. |
| 33 | ZOU H, HASTIE T. Regularization and variable selection via the elastic net[J]. J R Stat Soc B, 2005, 67(2): 301-320. |
| 34 | KRIZHEVSKY A, SUTSKEVER I, HINTON G E. Image net classification with deep convolutional neural networks[J]. Commun ACM, 2017, 60(6): 84-90. |
| 35 | ESTERHUIZEN J A, GOLDSMITH B R, LINIC S. Interpretable machine learning for knowledge generation in heterogeneous catalysis[J]. Nat Catal, 2022, 5(3): 175-184. |
| 36 | FUNG V, HU G, GANESH P, et al. Machine learned features from density of states for accurate adsorption energy prediction[J]. Nat Commun, 2021, 12(1): 88. |
| 37 | WANG S H, PILLAI H S, WANG S, et al. Infusing theory into deep learning for interpretable reactivity prediction[J]. Nat Commun, 2021, 12(1): 5288. |
| 38 | LAN T, AN Q. Discovering catalytic reaction networks using deep reinforcement learning from first-principles[J]. J Am Chem Soc, 2021, 143(40): 16804-16812. |
| 39 | MORET M, FRIEDRICH L, GRISONI F, et al. Generative molecular design in low data regimes[J]. Nat Mach Intell, 2020, 2(3): 171-180. |
| 40 | LI X, LI B, YANG Z, et al. A transferable machine-learning scheme from pure metals to alloys for predicting adsorption energies[J]. J Mater Chem A, 2022, 10(2): 872-880. |
| 41 | GAULTON A, BELLIS L J, BENTO A P, et al. ChEMBL: a large-scale bioactivity database for drug discovery[J]. Nucleic Acids Res, 2012, 40(D1): D1100-D1107. |
| 42 | LENSELINK E B, TEN DIJKE N, BONGERS B, et al. Beyond the hype: deep neural networks outperform established methods using a ChEMBL bioactivity benchmark set[J]. J Cheminf, 2017, 9(1): 1-14. |
| 43 | MA J, SHERIDAN R P, LIAW A, et al. Deep neural nets as a method for quantitative structure-activity relationships[J]. J Chem Inf Model, 2015, 55(2): 263-274. |
| 44 | UNTERTHINER T, MAYR A, KLAMBAUER G, et al. Multi-task deep networks for drug target prediction: Neural Information Processing System(NeurIPS)[C]. Montréal, CAN, 2014: 1-4. |
| 45 | BJERRUM E J. Smiles enumeration as data augmentation for neural network modeling of molecules[J]. arXiv preprint arXiv, 2017: 1703.07076. |
| 46 | SEGLER M H S, KOGEJ T, TYRCHAN C, et al. Generating focused molecule libraries for drug discovery with recurrent neural networks[J]. ACS Central Sci, 2018, 4(1): 120-131. |
| 47 | DUVENAUD D, MACLAURIN D, AGUILERA-IPARRAGUIRRE J, et al. Convolutional networks on graphs for learning molecular fingerprints[J]. arXiv preprint arXiv, 2015: 1509.09292. |
| 48 | COLEY C W, BARZILAY R, GREEN W H, et al. Convolutional embedding of attributed molecular graphs for physical property prediction[J]. J Chem Inf Model, 2017, 57(8): 1757-1772. |
| 49 | COREY E J, WIPKE W T. Computer assisted design of complex organic synthesis[J]. Science, 1969, 166: 178-192. |
| 50 | COREY E J, CRAMER R D, HOWE W J. Computer-assisted synthetic analysis for complex molecules. Methods and procedures for machine generation of synthetic intermediates[J]. J Am Chem Soc, 1972, 94(2): 440-459. |
| 51 | GELERNTER H, ROSE J R, CHEN C. Building and refining a knowledge base for synthetic organic chemistry via the methodology of inductive and deductive machine learning[J]. J Chem Inf Model, 1990, 30(4): 492-504. |
| 52 | LAW J, ZSOLDOS Z, SIMON A, et al. Route designer: a retrosynthetic analysis tool utilizing automated retrosynthetic rule generation[J]. J Chem Inf Model, 2009, 49(3): 593-602. |
| 53 | COLEY C W, ROGERS L, GREEN W H, et al. SCScore: synthetic complexity learned from a reaction corpus[J]. J Chem Inf Model, 2018, 58(2): 252-261. |
| 54 | SCHWALLER P, GAUDIN T, LÁNYI D, et al. “Found in translation”: predicting outcomes of complex organic chemistry reactions using neural sequence-to-sequence models[J]. Chem Sci, 2018, 9(28): 6091-6098. |
| 55 | WINTER R, MONTANARI F, NOÉ F, et al. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations[J]. Chem Sci, 2019, 10(6): 1692-1701. |
| 56 | SEGLER M H S, WALLER M P. Neural-symbolic machine learning for retrosynthesis and reaction prediction[J]. Chem-Eur J, 2017, 23(25): 5966-5971. |
| 57 | SEGLER M H S, PREUSS M, WALLER M P. Planning chemical syntheses with deep neural networks and symbolic AI[J]. Nature, 2018, 555(7698): 604-610. |
| 58 | NIROGI R V, BADANGE R, REBALLI V, et al. Design, synthesis and biological evaluation of novel benzopyran sulfonamide derivatives as 5-HT 6 receptor ligands[J]. Asian J Chem, 2015, 27(6): 2117-2124. |
| 59 | FUNES-ARDOIZ I, SCHOENEBECK F. Established and emerging computational tools to study homogeneous catalysis-from quantum mechanics to machine learning[J]. Chem, 2020, 6(8): 1904-1913. |
| 60 | HATTORI T, KITO S. Neural network as a tool for catalyst development[J]. Catal Today, 1995, 23(4): 347-355. |
| 61 | TIAN X, CHEN M. Descriptor selection for predicting interfacial thermal resistance by machine learning methods[J]. Sci Rep-UK, 2021, 11(1): 739. |
| 62 | GREELEY J, NØRSKOV J K. Combinatorial density functional theory-based screening of surface alloys for the oxygen reduction reaction[J]. J Phys Chem C, 2009, 113(12): 4932-4939. |
| 63 | MEDFORD A J, VOJVODIC A, HUMMELSHØJ J S, et al. From the Sabatier principle to a predictive theory of transition-metal heterogeneous catalysis[J]. J Catal, 2015, 328: 36-42. |
| 64 | TOYAO T, MAENO Z, TAKAKUSAGI S, et al. Machine learning for catalysis informatics: recent applications and prospects[J]. ACS Catal, 2020, 10(3): 2260-2297. |
| 65 | KIM E, HUANG K, SAUNDERS A, et al. Materials synthesis insights from scientific literature via text extraction and machine learning[J]. Chem Mater, 2017, 29(21): 9436-9444. |
| 66 | KIM E, HUANG K, TOMALA A, et al. Machine-learned and codified synthesis parameters of oxide materials[J]. Sci Data, 2017, 4(1): 170127. |
| 67 | TRAN K, ULISSI Z W. Active learning across intermetallics to guide discovery of electrocatalysts for CO2 reduction and H2 evolution[J]. Nat Catal, 2018, 1(9): 696-703. |
| 68 | TAKIGAWA I, SHIMIZU K I, TSUDA K, et al. Machine-learning prediction of the d-band center for metals and bimetals[J]. RSC Adv, 2016, 6(58): 52587-52595. |
| 69 | TOYAO T, SUZUKI K, KIKUCHI S, et al. Toward effective utilization of methane: machine learning prediction of adsorption energies on metal alloys[J]. J Phys Chem C, 2018, 122(15): 8315-8326. |
| 70 | JINNOUCHI R, ASAHI R. Predicting catalytic activity of nanoparticles by a DFT-aided machine-learning algorithm[J]. J Phys Chem Lett, 2017, 8(17): 4279-4283. |
| 71 | HUMMELSHØJ J S, ABILD-PEDERSEN F, STUDT F, et al. CatApp: a web application for surface chemistry and heterogeneous catalysis[J]. Angew Chem Int Ed, 2012, 51(1): 272-274. |
| 72 | WINTHER K T, HOFFMANN M J, BOES J R, et al. Catalysis-Hub.org, an open electronic structure database for surface reactions[J]. Sci Data, 2019, 6(1): 75. |
| 73 | CHANUSSOT L, DAS A, GOYAL S, et al. Open Catalyst 2020 (OC20) dataset and community challenges[J]. ACS Catal, 2021, 11(10): 6059-6072. |
| 74 | MADSEN J, LIU P, KLING J, et al. A deep learning approach to identify local structures in atomic-resolution transmission electron microscopy images[J]. Adv Theor Simul, 2018, 1(8): 1800037. |
| 75 | GHOSH K, STUKE A, TODOROVIĆ M, et al. Deep learning spectroscopy: neural networks for molecular excitation spectra[J]. Adv Sci, 2019, 6(9): 1801367. |
| 76 | LIU J, OSADCHY M, ASHTON L, et al. Deep convolutional neural networks for Raman spectrum recognition: a unified solution[J]. Analyst, 2017, 142(21): 4067-4074. |
| 77 | LAMOUREUX P S, WINTHER K T, TORRES J A G, et al. Machine learning for computational heterogeneous catalysis[J]. ChemCatChem, 2019, 11(16): 3581-3601. |
| 78 | PETERSON A A. Acceleration of saddle-point searches with machine learning[J]. J Chem Phys, 2016, 145(7): 074106. |
| [1] | 杨弘力, 王倩琪, 汪欢, 颜晗, 林芃晖, 宋金秋, 丁洋, 李善花, 李福男. 肉桂酰胺类化合物的结构优化、合成及其抗肝癌活性[J]. 应用化学, 2023, 40(2): 261-267. |
| [2] | 刘志浩, 谭晓晴, 梁永鹏, 夏超, 孟建新, 李风煜. 仿生视觉色度识别的浓度快速测定[J]. 应用化学, 2022, 39(1): 196-204. |
| [3] | 何冰,钟鑫鑫,冉凯,奉强,余登斌,韩涛,李仲辉,余洛汀. 硝基呋喃亚甲基哌啶类化合物的合成与抗结核活性[J]. 应用化学, 2020, 37(2): 134-143. |
| [4] | 惠康龙, 傅继澎, 高湉, 唐明学. 金属硫化物在可充电电池中的研究进展[J]. 应用化学, 2020, 37(12): 1384-1402. |
| [5] | 汤海峰,崔凤超,刘伦洋,李云琦. 基于配体与受体结构的酪氨酸酶抑制剂定量构效关系分析[J]. 应用化学, 2018, 35(7): 788-794. |
| [6] | 乐传俊, 王丽, 苏扬, 朱少萍. 甘油的化学转化研究概况[J]. 应用化学, 2014, 31(04): 367-376. |
| [7] | 于涛, 刘华沙, 王超群, 丁伟, 曲广淼. 烷基芳基磺酸钠对烷烃的乳化性能[J]. 应用化学, 2011, 28(05): 560-564. |
| [8] | 江银枝, 周俊, 梁大伟, 蒋玲波, 刘正江. 4, 6-二芳基-2-氨基嘧啶类衍生物的合成与生物活性[J]. 应用化学, 2011, 28(02): 209-213. |
| [9] | 周金梅, 王毅, 汤培平, 武小满, 林国栋, 张鸿斌. 甲烷在流态化催化剂床裂解生长多壁碳纳米管[J]. 应用化学, 2005, 22(2): 117-122. |
| [10] | 冯长君, 王超, 杨伟华. 分子树拓扑指数与羧酸化合物pKa值的定量构效关系[J]. 应用化学, 2004, 21(5): 469-474. |
| [11] | 冯长君, 堵锡华, 唐自强. 取代芳烃对发光菌、大型蚤、呆鲦鱼急性毒性的QSAR研究[J]. 应用化学, 2002, 19(11): 1037-1042. |
| [12] | 章文军, 许禄. HEPT类化合物的QSAR研究[J]. 应用化学, 2001, 18(9): 717-720. |
| [13] | 章文军, 徐禄. 定量结构-活性/性质相关性研究中变量选择方法的研究——遗传算法和几种传统算法的比较[J]. 应用化学, 2001, 18(3): 188-191. |
| [14] | 丁宇, 杨光富. B环取代的新型磺烷酮衍生物的合成及杀菌活性[J]. 应用化学, 2001, 18(10): 785-789. |
| [15] | 周嘉, 陈茹玉, 杨华铮, 杨秀凤, 杨光富. 硫(硒)代磷酸酯-膦酸酯类化合物除草活性的构效关系[J]. 应用化学, 1998, 0(3): 19-21. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||